Recent advances in genome-scale sequencing methods have resulted in a significant increase in our understanding of the biology of human cancers. When applied to pediatric central nervous system (CNS) tumors, these remarkable technological breakthroughs have facilitated the molecular characterization of multiple tumor types, provided new insights into the genetic basis of these cancers, and prompted innovative strategies that are changing the management paradigm in pediatric neuro-oncology. Genomic tests have begun to affect medical decision making in a number of ways, from delineating histopathologically similar tumor types into distinct molecular subgroups that correlate with clinical characteristics, to guiding the addition of novel therapeutic agents for patients with high-risk or poor-prognosis tumors, or alternatively, reducing treatment intensity for those with a favorable prognosis. Genomic sequencing has also had a significant impact on translational research strategies in pediatric CNS tumors, resulting in wide-ranging applications that have the potential to direct the rational preclinical screening of novel therapeutic agents, shed light on tumor heterogeneity and evolution, and highlight differences (or similarities) between pediatric and adult CNS tumors. Finally, in addition to allowing the identification of somatic (tumor-specific) mutations, the analysis of patient-matched constitutional (germline) DNA has facilitated the detection of pathogenic germline alterations in cancer genes in patients with CNS tumors, with critical implications for genetic counseling and tumor surveillance strategies for children with familial predisposition syndromes. As our understanding of the molecular landscape of pediatric CNS tumors continues to advance, innovative applications of genomic sequencing hold significant promise for further improving the care of children with these cancers.
Background
Tremendous advances have been achieved in DNA sequencing technologies since the early adoption of this method by Frederick Sanger in 1975.[1] Sequencing of the entire human genome was first achieved in 2003; this accomplishment required 13 years of work and $2.7 billion. By 2008, whole-exome sequencing (WES; the sequencing of all the protein-coding genes in a genome) of human cancers was first accomplished, at a cost of approximately $100,000 per case.[2] An early tumor exome study focused on glioblastoma multiforme (GBM), the most common malignant brain tumor in adults, unexpectedly identifying hotspot mutations in the IDH1 and IDH2 genes in a subset (< 10%) of GBMs that shared characteristic clinical and genetic features.[3,4] This discovery of recurrent mutations in a gene not previously linked to cancer became critical to the clinical and biologic understanding of pediatric and adult GBM; it provided prognostic data, facilitated the future molecular classification of GBM, and spurred research into epigenetic and metabolic aspects of both low- and high-grade gliomas.
More recently, high-throughput next-generation sequencing methods have incorporated automated and simultaneous sequencing of millions of short DNA or RNA fragments, combining this with advanced platforms for bioinformatics analysis, with the result that an entire genome can be interrogated in a matter of days at a cost of a few thousand dollars.[5] The specific applications of these methods are numerous: the sequence of nucleotides can be determined for an entire genome (whole-genome sequencing [WGS]), for an exome (WES), for a transcriptome (RNA sequencing [RNA-seq]), or for a targeted set of genes (mutation panels). Other common methods of detecting different types of alterations in tumor genomes include the use of array-based comparative genomic hybridization (aCGH) or copy number arrays to detect copy number variations (CNVs)-eg, amplifications or deletions, or loss of heterozygosity-and the use of techniques such as methylation arrays to perform epigenetic analysis. These diverse methods now allow comprehensive evaluation of the various aberrant molecular pathways underlying tumorigenesis in individual patients or specific tumor types, thereby improving our understanding of the biology of these cancers and allowing for innovative approaches to their clinical management. This review will discuss examples of clinical applications of tumor and germline genomic testing for children with central nervous system (CNS) tumors, as well as promising investigative strategies.
Incorporation of Molecular Characteristics Into Pathologic Diagnosis
As with other cancer types, the incorporation of molecular data into previously established histopathologic criteria has led to a remarkable refinement in the diagnosis of childhood CNS tumors. A classic example is the group of pediatric embryonal CNS tumors, once considered identical small round blue cell tumors arising in various neuroanatomic locations but now recognized as distinct diagnostic entities defined by unique molecular and clinical signatures: (a) atypical teratoid/rhabdoid tumor (AT/RT), an aggressive tumor that occurs most frequently in young children; (b) supratentorial primitive neuroectodermal tumors (sPNETs); and (c) medulloblastoma, which occurs exclusively in the posterior fossa.
AT/RT was first distinguished from medulloblastoma and sPNET in the 1980s on the basis of the presence of rhabdoid cells-cells with a distinctive eccentric round nucleus, a prominent nucleolus, and a plump cell body. However, this tumor remained diagnostically challenging, since rhabdoid cells can vary in size and features such that rhabdoid tumors often have a histopathologic appearance similar to those of sPNET and medulloblastoma.[6] The discovery of deletions and inactivating mutations in SMARCB1/INI1 enabled the molecular diagnosis of AT/RT. These inactivating alterations in SMARCB1/INI1, and to a lesser extent in SMARCA4,[7] impair the coding of components of the SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex[8] and result in a corresponding loss of protein detectable by immunohistochemical staining.[9] Remarkably, AT/RT has been found to be an otherwise genomically silent tumor (nearly devoid of other recurrent genomic alterations) despite its aggressive nature,[10,11] and strategies that target perturbations of epigenetic modification due to INI1 deficiency have generated significant interest.[12] Interestingly, studies suggest that sPNETs without rhabdoid cells on histopathology tend to behave more aggressively and have a poorer prognosis when they exhibit a loss of INI protein expression.[13]
CNS sPNET has historically been distinguished from medulloblastoma by its distinct neuroanatomic location, but a biologic understanding of this tumor is emerging.[14] The current World Health Organization (WHO) classification schema[15] identifies the following histopathologic variants of sPNET: embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma. However, recent genomic analyses have revealed shared molecular aberrations across the histopathologic variants; fluorescence in situ hybridization (FISH) analysis has demonstrated 19q13.42 amplifications involving the C19MC cluster across all three variants; gene expression and methylation profiles were also similar, suggestive of a single molecular entity despite variations in histologic appearance.[16,17] This group of tumors has been labeled Group 1, or embryonal tumor with multilayered rosettes (ETMR); tumors in this group have been associated with similar clinical features, including young patient age (< 4 years) and poor outcome (survival of 12 to 36 months).[17] Gene expression profiling and clinical data suggest that sPNETs comprise two additional subgroups: Group 2 tumors, characterized by oligoneuronal gene upregulation and a slightly less poor prognosis (median survival, 1.8 years), and Group 3 tumors, characterized by high expression of epithelial and mesenchymal differentiation genes and better clinical outcomes (median survival, 4.3 years). While the molecular subtyping of sPNETs has not yet altered standard treatment approaches to this high-risk tumor, the groups can be reliably delineated with readily available immunostains (LIN28 and OLIG2), and this will aid in the development and analysis of future clinical trials based upon molecular classification,[14,18] an approach that is now becoming standard in the treatment of medulloblastoma (see next section).
Medulloblastoma: Biologic and Prognostic Management
The rapid evolution of pediatric medulloblastoma management represents arguably the most advanced utilization of genomic data in pediatric neuro-oncology. Medulloblastoma, the most common malignant brain tumor in children, is an invasive, midline neuro-epithelial neoplasm that arises from the cerebellum. Historically, these tumors were considered a single disease and were treated in a uniform manner, based on clinical characteristics such as patient age, presence of metastatic disease, and extent of tumor resection. Initial attempts to classify medulloblastoma employed histologic subtypes (eg, classic, nodular/desmoplastic, large-cell/anaplastic) that were associated with a limited degree of prognostic significance. An early strategy for the molecular risk stratification of patients was based on the observed association between poor survival and the presence of tumor MYC or MYCN amplification.[19,20] Detection of MYC and MYCN amplification is readily accomplished using FISH or aCGH, and this marker was subsequently incorporated into risk-stratified large-scale clinical trials.[21,22]
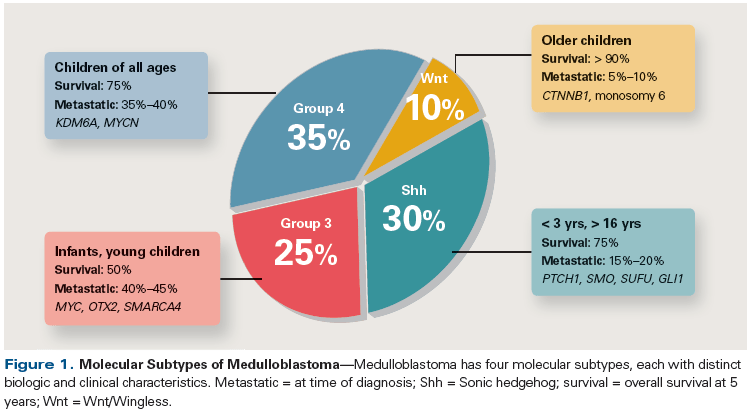
The clinical approach to patients with medulloblastoma shifted further following landmark studies that characterized the molecular (genetic, transcriptomic, and epigenomic) landscape of this tumor, leading to the classification of medulloblastoma into four biologically distinct and clinically relevant subgroups: tumors defined by activation of the Wnt/Wingless developmental pathway (Wnt subtype), those defined by activation of the Sonic hedgehog pathway (Shh subtype), Group 3, and Group 4 (Figure 1). The recognition of these molecular subtypes has created an opportunity to reduce treatment intensity and toxicity for subgroups recognized as having excellent long-term survival (Wnt subtype), while identifying new targets for novel and targeted therapies. These four subgroups were first described using tumor expression profiling and copy number analyses of large medulloblastoma cohorts[23-25]; subsequent studies utilizing genomic sequencing and methylation arrays have confirmed the classification into these molecular subgroups.[26-28] The optimal molecular test for routine clinical use has yet to be definitively established (possibilities include immunohistochemistry, next-generation sequencing, methylation arrays, and other methods); nonetheless, subgroup classification of medulloblastoma has been rapidly incorporated into multiple large-scale clinical trials in order to assign investigational therapies based on risk status and the molecular features of each patient’s tumor.
Wnt subtype medulloblastoma is characterized by dysregulation in canonical Wnt pathway signaling, frequently due to somatic CTNNB1 (beta-catenin) mutations. This subtype demonstrates the opportunity for genomically driven prognostic information to impact the delivery of risk-adapted adjuvant therapy for patients with childhood CNS tumors. Although only 10% of childhood medulloblastomas fall into the Wnt category, this population of patients exhibits an excellent overall survival (OS) rate of greater than 95%.[21,29] The favorable outcome and response of these tumors with conventional treatment have led to ongoing clinical trials investigating the feasibility of reducing the craniospinal irradiation dose in an attempt to limit treatment-related toxicity in this patient cohort.
Shh subtype medulloblastoma is associated with mutations in genes that are part of the Shh signaling pathway, including SMO, PTCH, SUFU, and GLI2. This subtype has been associated with an intermediate prognosis (OS, 60% to 80%)[21,29] and occurs most frequently in two age groups: very young children and adults. Importantly, the distribution of specific gene mutations correlates with patient age group, with SUFU mutations enriched in the cohort of very young children and SMO mutations most often seen in adult patients, while tumors that harbor PTCH mutations span both groups. This variability in gene mutations has significant implications for use of inhibitors that target SMO, such as vismodegib,[30] as tumors that harbor PTCH or SMO mutations (more common in adult patients) appear more responsive than SUFU-mutated tumors.[31] A subset of tumors in the Shh subgroup have a particularly poor prognosis (compared with other Shh tumors) and have been found to harbor TP53 mutations, many of which are germline rather than somatic events-eg, the germline mutation of TP53 that causes the Li-Fraumeni cancer predisposition syndrome, which, when identified, indicates a need for genetic counseling and appropriate cancer surveillance for the affected patient and his or her family.[32,33]
Of the medulloblastoma subtypes, the biology of Group 3 and Group 4 tumors is least understood; tumors of these two subtypes account for more than half of medulloblastomas and are associated with an inferior prognosis (OS of 40%–60% and ~75%, respectively).[21,29] Recent work has revealed the activation of oncogenes GFI1 and GFI1B in one-third of Group 3 medulloblastomas and in 5% to 10% of Group 4 medulloblastoma patients.[34] This oncogenic activation is effected not by mechanisms directly involving the target gene, but instead by an indirect mechanism termed “enhancer hijacking.” GFI1 and GFI1B are usually in regions of transcriptionally silent chromatin. Structural variants caused by diverse mechanisms, such as deletion, inversion, tandem duplication, and translocation, result in genomic rearrangements that place the coding sequences of GFI1 or GFI1B proximal to active enhancers of transcription, thus activating these proto-oncogenes. Specific chemotherapeutic agents, such as pemetrexed and gemcitabine, have been identified through high-throughput drug screening strategies as active agents against Group 3 medulloblastoma[35]; however, additional information regarding specific targets for therapy in these subgroups is still needed.
Single- and Multiple-Pathway Diseases
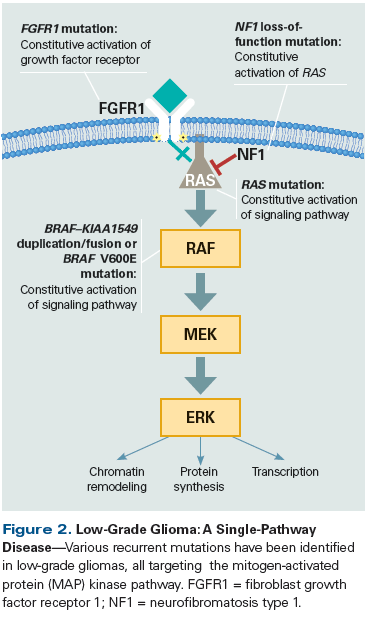
Comprehensive genomic analyses of CNS tumors have revealed that certain tumor types have alterations exclusively within a single molecular pathway, while others have more complex genomes and diverse alterations affecting multiple pathways and biologic processes. Pilocytic astrocytoma (WHO grade I astrocytoma), the most common CNS tumor in children, is an example of a single-pathway disease (Figure 2). More than 80% of these tumors exhibit a fusion of the BRAF and KIAA1549 genes, with the resulting loss of the autoregulatory domain of BRAF, which causes its constitutive activation.[36,37] In pilocytic astrocytomas that lack this characteristic BRAF fusion, a variety of other genetic alterations have been found that all activate the mitogen-activated protein (MAP) kinase pathway, including other BRAF fusions, such as FAM131B–BRAF and SRGAP3–RAF1,[38,39] point mutations in BRAF (most commonly the V600E missense mutation seen in melanoma and other cancers), upstream pathway mutations in FGFR1 (which causes constitutive activation of the fibroblast growth factor receptor kinase),[40] and mutations in KRAS.[40,41] Both germline and somatic loss-of-function mutations in tumor suppressor genes such as NF1 can also result in prolonged activation of the MAP kinase pathway. Other examples of tumor types predominantly dependent on disruption of a single molecular pathway include: ganglioglioma (BRAF V600E mutation, KIAA1549–BRAF fusion),[42] angiocentric glioma (MYB–QKI fusion),[42,43] adamantinomatous subtype craniopharyngioma (CTNNB1 mutation), and papillary subtype craniopharyngioma (BRAF V600E mutation).[44,45]
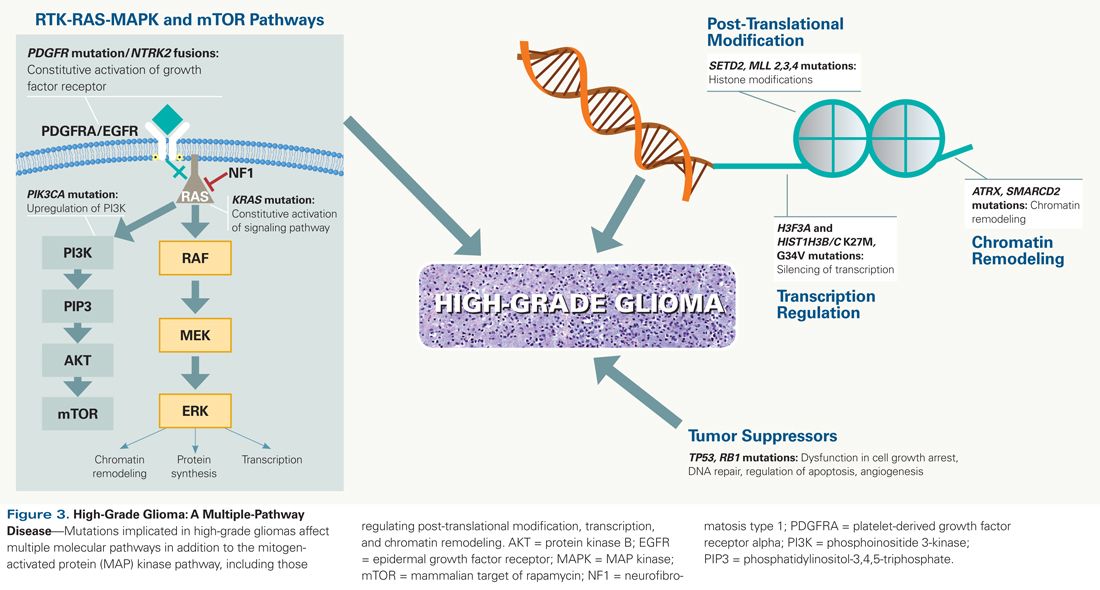
In contrast to single-pathway tumors, high-grade pediatric gliomas have been found to harbor a wide variety of genetic aberrations affecting multiple signaling pathways and cancer-related biological functions (Figure 3). Notably, many of these are characterized by mutations in genes involved in epigenetic regulation, including histone gene mutations (H3F3A and HIST1H3B), genes related to chromatin remodeling (ATRX, ARID1B, SMARCA4), and genes implicated in post-translational modification of histones and chromatin (MLL2/KMT2D, MLL3/KMT2C, SETD2).[46] In all, mutations affecting epigenetic regulation are found in more than 90% of diffuse intrinsic pontine gliomas, more than two-thirds of midline non-brainstem high-grade gliomas, and nearly half of hemispheric high-grade gliomas. In addition to these mutations in epigenetic genes, alterations are also detected in a number of other cancer genes and pathways, including TP53 and RB1, which act as gatekeepers for cell-cycle progression and as caretakers of genome maintenance. Finally, mutations, amplifications, and fusions causing activation of the MAP kinase and phosphoinositide 3-kinase (PI3K) signaling pathways (BRAF V600E, NF1, PIK3CA, and ACVR1 mutations; PDGFRA mutation and amplification; NTRK2 fusions) occur in more than two-thirds of high-grade pediatric gliomas.
Cancer Predisposition Syndromes in Children With CNS Tumors
Genome-scale methods for analyzing constitutional DNA (such as WES) have emerged as a promising diagnostic strategy for identifying familial predisposition syndromes in pediatric cancer patients. In recent exome and genome sequencing studies of pediatric oncology patients with diverse diagnoses, a pathogenic or likely pathogenic germline cancer gene variant was detected in 8% to 10% of patients, highlighting the importance of considering the possibility of germline mutations in these children.[47-49] Although most clinical tumor sequencing is currently performed without the parallel analysis of a matched normal tissue (blood) sample, recent studies have emphasized the importance of such paired analyses to both improve identification of somatic mutations and distinguish germline variants from somatic mutations.[50]
Although the prevalence of germline mutations underlying familial cancer syndromes in the population of children with CNS tumors has yet to be determined, several tumor types are well-known sequelae of familial predisposition syndromes. For example, analysis of two large cohorts of patients with AT/RT identified a SMARCB1 germline pathogenic alteration in 17/40 (43%) and 23/65 (35%), signifying a predisposition for the development of other rhabdoid tumors.[51,52] Similarly, patients with Li-Fraumeni syndrome, characterized by pathogenic germline TP53 variants, are at high risk for high-grade gliomas, choroid plexus carcinoma, and medulloblastoma,[33,53] while patients with neurofibromatosis type I are at particular risk for low-grade gliomas, with ~15% of patients developing this neoplasm in the first 2 decades of life.[54,55] Screening guidelines and specialized clinics that care for this patient population (and other affected family members) are increasing in frequency, highlighting the importance of having genetic counseling and cancer genetics expertise available at institutions that perform this type of genomic testing.[33]
Beyond the Genome: Transcriptome and Epigenome Sequencing
Although WGS and WES focus on detection of genetic alterations in genomic DNA, other sequencing modalities offer the potential to provide complementary data of biological and clinical importance for cancer patients. Transcriptome sequencing has played a significant role in the discovery of novel gene fusions that occur in multiple cancer types, including specific pediatric CNS tumors, such as supratentorial ependymomas. Recent work by Parker et al uncovered a novel translocation in these tumors resulting in fusion of the RELA gene, the principal effector of canonical nuclear factor–kappa B signaling, and C11orf95, a poorly characterized gene. This translocation has been found in more than two-thirds of supratentorial ependymomas, but never in posterior fossa or spinal ependymomas,[56] and was demonstrated to be tumorigenic when aberrantly expressed in pluripotent neural stem cells. In another study, more than half of supratentorial ependymomas negative for the RELA fusion were found instead to harbor novel translocations involving YAP1 and a variety of partner genes. As was observed for RELA, the YAP1 translocations were also unique to supratentorial ependymomas and were not found in posterior fossa or spinal tumors.[57]
Posterior fossa ependymomas exhibit very different molecular signatures. Gene expression profiling has identified two distinct types of posterior fossa ependymomas. Group A tumors tend to occur in younger children (median age, 2.5 years); develop in a lateral location (67% of patients); and have relatively poor outcomes, with 5-year progression-free survival (PFS) and OS rates of 47% and 59%, respectively. In contrast, group B tumors, which cluster with the transcriptional profile of spinal ependymomas, occur predominantly in older patients (median age, 20 years); develop in a central location (95% of patients); and have a relatively better prognosis, with 5-year PFS and OS rates of 79% and 95%, respectively.[58]
The use of complementary genomic modalities may be particularly important for pediatric cancers, including CNS tumors, compared with adult tumors, given the relative paucity of recurrent somatic mutations and CNVs in pediatric cases. Notably, epigenetic changes (changes that result in alterations in gene expression without a change in the actual DNA or RNA sequence) have been found to play a major role in tumorigenesis for certain pediatric CNS tumors. Methylation profiling is a method of studying the regulation of gene expression through DNA methylation, which results in the silencing (loss of expression) of targeted genes. Methylation profiling of ependymomas confirmed the same subgroups revealed earlier by transcriptional profiling. Group A posterior fossa ependymomas, in comparison with group B ependymomas, were found to have significantly more genes with hypermethylation in the CpG promoter region and thus more genes that were transcriptionally silenced.[59] Interestingly, the genes silenced in group A ependymomas are required for differentiation of embryonic stem cells. These genes are known to be silenced by polycomb repressive complex 2 and have been demonstrated in other studies to frequently undergo cancer-specific CpG methylation.[60]
From Bench to Bedside: CNS Tumor Models and Preclinical Testing
TO PUT THAT INTO CONTEXT
[[{"type":"media","view_mode":"media_crop","fid":"48691","attributes":{"alt":"","class":"media-image","id":"media_crop_7216928302696","media_crop_h":"0","media_crop_image_style":"-1","media_crop_instance":"5843","media_crop_rotate":"0","media_crop_scale_h":"0","media_crop_scale_w":"0","media_crop_w":"0","media_crop_x":"0","media_crop_y":"0","style":"height: 165px; width: 144px;","title":" ","typeof":"foaf:Image"}}]]
Scott L. Pomeroy, MD, PhD
Boston Children’s Hospital
Boston, MassachusettsCancer genomics has revolutionized our basic understanding of childhood brain tumors. Genomic profiling has delineated molecular subgroups of tumors with unique genetic lesions and different clinical outcomes despite similar histopathology. This has made a more accurate assessment of prognosis possible, resulting in the appropriate intensification of conventional treatments for high-risk patients and the reduction of treatment for those predicted to have a favorable outcome. Genomic profiling has also led to the introduction of novel therapies that target molecular mechanisms.How Has Genomic Profiling Affected the Classification of Childhood Brain Tumors? Embryonal tumors have been delineated into atypical teratoid/rhabdoid tumors, with characteristic inactivating lesions of SMARCB1/INI1 and very poor prognosis; medulloblastomas; and supratentorial primitive neuroectodermal tumors (sPNETs). Four principal subgroups of medulloblastoma have been identified: one defined by activation of the Sonic hedgehog (Shh) pathway (Shh subgroup); one defined by activation of the Wnt/Wingless pathway (Wnt subgroup); and two with less certain mechanisms of oncogenesis, including Group 3, dominated by MYC expression, and Group 4.What Are Some of the Clinical Implications of the New Classification? Tumors in the Wnt subgroup have an excellent prognosis, allowing for the reduction of neurotoxic treatment with radiation and chemotherapy. Clinical trials with targeted therapies are in progress for tumors in the Shh subgroup. The sPNETs have proven to be a highly heterogeneous group of tumors, including a subgroup with high LIN28 expression (which has a very poor prognosis) and other subgroups that more closely resemble gliomas and other tumor types. Low-grade gliomas are dominated by molecular lesions that activate the mitogen-activated protein (MAP) kinase pathway and that are suitable targets for small-molecule therapies, whereas high-grade gliomas have mutations that alter epigenetic regulation of gene transcription, which makes targeting them with small-molecule therapies more challenging. Heterogeneity within tumors has also been revealed, including molecularly unique subclones that underlie metastasis. Collectively, these discoveries have redefined tumor classification and have ushered in the era of precision medicine for childhood brain tumors.
Although genomic analyses of pediatric CNS tumors have revealed a number of mutations in known cancer genes and pathways, the vast majority of alterations identified to date are of unclear biologic significance. In order to further evaluate the functional and clinical significance of both known and novel mutations, significant efforts have focused on the development of pediatric CNS tumor models that can be utilized for biologic studies and preclinical evaluation of investigational agents. Orthotopic xenograft mouse models have been developed for the major types of pediatric brain tumors-such as medulloblastoma, sPNET, and ependymoma-by direct injection of patient tumor into the corresponding anatomic location in the mouse brain, and have been demonstrated to faithfully retain specific genetic aberrations and gene expression signatures of the original tumor.[61-63] The genomic characterization of orthotopic xenografts will be increasingly important in order to facilitate the evaluation of therapies targeting specific genetic alterations and molecular subtypes of pediatric CNS tumors. After drugs of interest are identified by in vitro testing using pediatric CNS tumor cell lines, they can then be tested in vivo utilizing mouse xenografts, which better mimic the tumor’s microenvironment. In addition to studying the effects of novel agents on tumor cell proliferation, tumor size, and mouse survival, studies using RNA sequencing can be performed to assess targeted gene expression of related targets.
A recent example of this strategy is the demonstration of the potential efficacy of panobinostat, a histone deacetylase inhibitor, for the treatment of diffuse intrinsic pontine glioma (DIPG).[64] During an initial in vitro screen using DIPG cell lines, panobinostat was found to be effective at reducing both the proliferation and survival of tumor cells. In vivo testing in xenograft models then revealed decreased tumor size and improved survival in mouse xenografts treated with panobinostat, compared with vehicle (dimethylsulfoxide) controls. In addition, RNA sequencing demonstrated normalization of the H3 K27M gene signature in panobinostat-treated tumors-an epigenetic change caused by the histone mutation known to underlie a significant proportion of aggressive midline high-grade gliomas.[64] Thus, sequencing and gene expression studies are being used to effectively guide the identification and testing of therapeutic targets and agents against specific pathways.
Tumor Heterogeneity
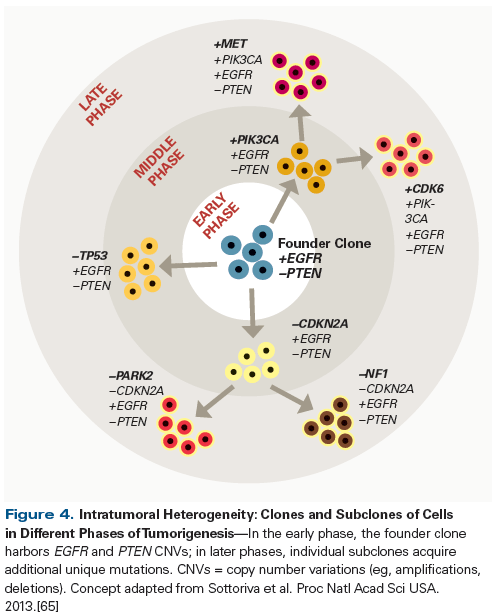
Studies have shown that a single tumor can contain multiple clones of tumor cells, with varying mutational signatures. In one study involving patients with GBM, multiple samples were obtained from each tumor during debulking surgery, then subjected to genome-wide copy number analysis (Figure 4).[65] One set of CNVs were found to be common to all the clones in a tumor, in a core set of genes classically described as being associated with GBM-such as EGFR, PDGFRA, and PTEN. A second set of CNVs were found to be shared by some-but not all-the clones in a tumor, while a third set were unique to individual clones.[65] Recently, single-cell RNA sequencing techniques demonstrated significant intratumoral heterogeneity in the transcriptional programs of primary GBM samples, despite the retention of chromosome-level alterations. These findings provide an additional biologic basis for the refractoriness of GBM to conventional therapies, but also an opportunity to identify new potential targets in molecular signatures associated with cell proliferation and treatment resistance.[66] Similar findings have been observed in other non-CNS tumors as well.[67-69] This suggests a clonal evolution model (see Figure 4), in which a set of driver mutations creates a founder clone of tumor cells in the early phase of tumorigenesis. This is followed by a middle phase, in which subclones are created by the acquisition of additional mutations. In later phases, additional unique mutations are acquired by some cells within a subclone. In this same study, different samples from the same tumor were found to be of different GBM subtypes, based on gene expression profiling.[65] The mechanism of the observed intratumoral heterogeneity remains an area of active research and debate: some studies suggest that this clonal evolution of cells is driven by Darwinian selection, in which tumor cells from one or multiple clones harboring mutations that confer fitness and greater resistance to therapy are selected to prevail over cells without those advantages.[67,70] On the other hand, the degree of genetic diversity and the finding that no single clone seems to predominate in a tumor suggest a non-Darwinian model of clonal evolution.[71] Irrespective of the mechanism of clonal evolution, the genomic diversity of the different clones within the same tumor has potentially significant implications for the selection of therapy targeting these aberrant molecular pathways. It is not yet clear how often the additional genetic aberrations acquired by new subclones are driver events that further promote tumorigenesis and change treatment resistance. These additional mutations could instead simply be passenger events that occur incidentally but do not significantly change the biology or behavior of the tumor.[2] Even when clones have unique driver mutations, phenotypically these mutations may converge on the same cellular pathways, making them vulnerable to the same targeted therapies.[70] However, if different cellular pathways/mechanisms are co-opted by the driver mutations specific to each clone, a multimodal approach with combinations of targeted agents or other treatment modalities, such as immunotherapy, might be more effective in ensuring the eradication of those clones and subclones.
Molecular heterogeneity has also been demonstrated between a primary CNS tumor and the metastases occurring in the same patient. In a recent study of medulloblastoma utilizing mouse xenografts and human samples, it was found that while the metastases from a patient were very similar genomically, they diverged significantly from the primary tumor, harboring only one of its subclones.[72] Significant genetic diversity has also been demonstrated between the diagnostic pre-therapy tumor and post-therapy recurrent tumor in medulloblastoma. WGS of paired samples of pre- and post-therapy tumor samples from children with medulloblastoma revealed that of the patients with targetable mutations, less than half retained the same mutation at relapse, more than half had a complete switch in actionable targets, and one-third gained new targetable mutations.[73] As with intratumoral molecular heterogeneity, the significance of the mutational heterogeneity between the primary tumor and its metastases requires further study to determine whether the mutations unique to the metastatic sites are driver events that change biology and treatment response to targeted therapies. If that proves to be the case, this genomic heterogeneity of different tumor sites in a patient is very significant for upfront therapy for a newly diagnosed patient, mandating a goal of optimizing therapy to eradicate resistant subclones and prevent tumor recurrence. Further, the molecular divergence of metastases from the primary tumor would make it imperative to not base clinical decisions at the time of recurrence on the genomic characteristics of the original tumor; it would also necessitate that clinical trials of molecularly targeted agents for recurrent solid CNS and non-CNS tumors mandate biopsy of the recurrent tumor to be eligible.
Future Directions
The recent pace of discoveries regarding the genetics and biology of pediatric CNS tumors is unprecedented, facilitating the development of genomically informed treatment plans to improve outcomes for future children with CNS tumors. Given the trajectory of advances in genomic technology (and their clinical application), it is entirely possible that in the near future, paired tumor and germline genome-scale analyses of children with CNS tumors will be performed in real time in their doctors’ offices, informing both the diagnosis and upfront management decisions. The diagnoses of certain histologically and biologically ill-defined tumor types, such as sPNETs, are likely to be completely revised on the basis of molecular data, with treatment being based on their molecular signatures. For other histopathologically well-characterized tumors, molecular subtypes have been identified that have distinct biologic characteristics and clinical features. These subtypes are likely to increasingly influence treatment decisions, allowing reduction in treatment intensity for certain tumor types with a good prognosis, while mandating intensification of treatment and the use of novel therapies such as molecularly targeted agents for tumor types with a relatively poor prognosis. Such treatment strategies are already being tested for medulloblastoma (ClinicalTrials.gov identifier: NCT01878617) and are likely to be incorporated in clinical trials for other brain tumor types, including ependymomas and high-grade gliomas, in the near future. The diagnostic yield of tumor genomic testing should continue to improve with the use of integrated modalities of genome-scale genetic testing (eg, RNA-seq and WES), facilitating improved identification of key molecular targets. It is hoped that these methods will improve outcomes, as well as reduce the side effects of current therapies for children with CNS tumors. Upcoming genomically guided clinical trials, such as the National Cancer Institute Pediatric Molecular Analysis for Therapy Choice (MATCH) study, will increasingly utilize genomic methods to evaluate the use of molecularly targeted agents, chosen based on the specific genetic aberration(s) identified in each patient’s tumor. We are rapidly entering an era of precision oncology in which the diagnosis, classification, and treatment of pediatric CNS tumors is informed by their molecular and genomic characteristics.
Financial Disclosure: The authors have no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
References:
1. Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441-8.
2. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546-58.
3. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807-12.
4. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765-73.
5. Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413-35.
6. Rorke LB, Packer RJ, Biegel JA. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: definition of an entity. J Neurosurg. 1996;85:56-65.
7. Fruhwald MC, Biegel JA, Bourdeaut F, et al. Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro Oncol. 2016 Jan 10. [Epub ahead of print]
8. Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203-6.
9. Judkins AR, Mauger J, Ht A, et al. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol. 2004;28:644-50.
10. Kieran MW, Roberts CW, Chi SN, et al. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatr Blood Cancer. 2012;59:1155-7.
11. Hasselblatt M, Isken S, Linge A, et al. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52:185-90.
12. Wetmore C, Boyett J, Li S, et al. Alisertib is active as single agent in recurrent atypical teratoid rhabdoid tumors in 4 children. Neuro Oncol. 2015;17:882-8.
13. Miller S, Ward JH, Rogers HA, et al. Loss of INI1 protein expression defines a subgroup of aggressive central nervous system primitive neuroectodermal tumors. Brain Pathol. 2013;23:19-27.
14. Picard D, Miller S, Hawkins CE, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 2012;13:838-48.
15. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (editors). WHO classification of tumours of the central nervous system. Lyon, France: IARC; 2007.
16. Spence T, Sin-Chan P, Picard D, et al. CNS-PNETs with C19MC amplification and/or LIN28 expression comprise a distinct histogenetic diagnostic and therapeutic entity. Acta Neuropathol. 2014;128:291-303.
17. Korshunov A, Sturm D, Ryzhova M, et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol. 2014;128:279-89.
18. Korshunov A, Ryzhova M, Jones DT, et al. LIN28A immunoreactivity is a potent diagnostic marker of embryonal tumor with multilayered rosettes (ETMR). Acta Neuropathol. 2012;124:875-81.
19. Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627-36.
20. Aldosari N, Bigner SH, Burger PC, et al. MYCC and MYCN oncogene amplification in medulloblastoma. A fluorescence in situ hybridization study on paraffin sections from the Children’s Oncology Group. Arch Pathol Lab Med. 2002;126:540-4.
21. Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813-20.
22. Strother D, Ashley D, Kellie SJ, et al. Feasibility of four consecutive high-dose chemotherapy cycles with stem-cell rescue for patients with newly diagnosed medulloblastoma or supratentorial primitive neuroectodermal tumor after craniospinal radiotherapy: results of a collaborative study. J Clin Oncol. 2001;19:2696-704.
23. Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408-14.
24. Cho YJ, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424-30.
25. Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924-31.
26. Robinson G, Parker M, Kranenburg TA, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43-8.
27. Pugh TJ, Weeraratne SD, Archer TC, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106-10.
28. Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465-72.
29. Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473-84.
30. Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173-8.
31. Kool M, Jones DT, Jager N, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393-405.
32. Rausch T, Jones DT, Zapatka M, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59-71.
33. Villani A, Tabori U, Schiffman J, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol. 2011;12:559-67.
34. Northcott PA, Lee C, Zichner T, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428-34.
35. Morfouace M, Shelat A, Jacus M, et al. Pemetrexed and gemcitabine as combination therapy for the treatment of Group 3 medulloblastoma. Cancer Cell. 2014;25:516-29.
36. Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673-7.
37. Pfister S, Janzarik WG, Remke M, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739-49.
38. Cin H, Meyer C, Herr R, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121:763-74.
39. Jones DT, Kocialkowski S, Liu L, et al. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene. 2009;28:2119-23.
40. Jones DT, Hutter B, Jager N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927-32.
41. Raabe E, Kieran MW, Cohen KJ. New strategies in pediatric gliomas: molecular advances in pediatric low-grade gliomas as a model. Clin Cancer Res. 2013;19:4553-8.
42. Qaddoumi I, Orisme W, Wen J, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016 Jan 25. [Epub ahead of print]
43. Bandopadhayay P, Ramkissoon LA, Jain P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016 Feb 1. [Epub ahead of print]
44. Brastianos PK, Taylor-Weiner A, Manley PE, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet. 2014;46:161-5.
45. Larkin SJ, Preda V, Karavitaki N, et al. BRAF V600E mutations are characteristic for papillary craniopharyngioma and may coexist with CTNNB1-mutated adamantinomatous craniopharyngioma. Acta Neuropathol. 2014;127:927-9.
46. Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444-50.
47. Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016 Jan 28. [Epub ahead of print]
48. Mody RJ, Wu YM, Lonigro RJ, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314:913-25.
49. Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336-46.
50. Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53.
51. Eaton KW, Tooke LS, Wainwright LM, et al. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer. 2011;56:7-15.
52. Bourdeaut F, Lequin D, Brugieres L, et al. Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res. 2011;17:31-8.
53. Ballinger ML, Mitchell G, Thomas DM. Surveillance recommendations for patients with germline TP53 mutations. Curr Opin Oncol. 2015;27:332-7.
54. Hernaiz Driever P, von Hornstein S, Pietsch T, et al. Natural history and management of low-grade glioma in NF-1 children. J Neurooncol. 2010;100:199-207.
55. Cnossen MH, de Goede-Bolder A, van den Broek KM, et al. A prospective 10 year follow up study of patients with neurofibromatosis type 1. Arch Dis Child. 1998;78:408-12.
56. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451-5.
57. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728-43.
58. Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143-57.
59. Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445-50.
60. Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237-42.
61. Liu Z, Zhao X, Wang Y, et al. A patient tumor-derived orthotopic xenograft mouse model replicating the group 3 supratentorial primitive neuroectodermal tumor in children. Neuro Oncol. 2014;16:787-99.
62. Zhao X, Liu Z, Yu L, et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol. 2012;14:574-83.
63. Yu L, Baxter PA, Voicu H, et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 2010;12:580-94.
64. Grasso CS, Tang Y, Truffaux N, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21:555-9.
65. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009-14.
66. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396-401.
67. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-92.
68. Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114-7.
69. Navin N, Krasnitz A, Rodgers L, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68-80.
70. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338-45.
71. Ling S, Hu Z, Yang Z, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci USA. 2015;112:E6496-E6505.
72. Wu X, Northcott PA, Dubuc A, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529-33.
73. Morrissy AS, Garzia L, Shih DJ, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529:351-7.