Molecular Profiling for Cytotoxics and Targeted Agents: Ready for Prime Time?
Lung cancer is the leading cause of cancer-related mortality. Improved understanding in the molecular biology and genetics of lung cancer has resulted in the identification of individual genes, gene expression profiles, and molecular pathways that may be useful for clinical management decisions.
Table of Contents
A Word From the Editor
Introduction
Prognostic Markers in Patients with NSCLC: ERCC1 and RRM1 as Markers of Improved Prognosis in NSCLC
Oligonucleotide-based Gene Expression Signatures of Improved Survival in Resected NSCLC Patients
Serum Proteomic Profiles as Prognostic Marker of Improved Prognosis in NSCLC
Markers That Predict for Therapeutic Efficacy
ERCCI as a Predictor for Platinum Resistance in Advanced NSCLC
RRMI as a Predictor of Gemcitabine Efficacy
BRCA1 as a Predictor of Chemotherapy Efficacy
Predictors of Improved Response or Survival with EGFR Tyrosine Kinase Inhibitors
Oligonucleotide-based Gene Expression Profiles as Predictors of Response in NSCLC Patients
Individualizing Treatment for Advanced Stage NSCLC Using ERCC1 and RRM1
Conclusions
References
CONTINUING MEDICAL EDUCATION Activity Release Date: December 15, 2008 Activity Expiration Date: December 15, 2009 About the Activity This activity is based on a brief article developed as part of the E-Update Series and posted on the Web. It was developed from an identified educational need for information about practical management issues in the practice of medical, surgical, and radiation oncology. This activity has been developed and approved under the direction of CME LLC. Activity Learning Objectives After reading this article, participants should be able to: 1. Demonstrate an understanding of the appropriate role of surgery in the metastatic setting: • Role of tackling the unexplored mediastinum in patients who have undergone resection • Role of surgery after chemoradiation in stage IIIA NSCLC 2. Apply into practice the role of radiation therapy post resection in the treatment of N2 NSCLC: •PORT = Post-Operative Radiation Therapy •STARBOARD = Sidestepping Trans-Thoracic Adjuvant Radiation Therapy Because One is Averse to Radiation Damage 3. Analyze the efficacy of molecular profiling for cytotoxics and targeted agents 4. Appraise the use of induction therapy versus adjuvant therapy 5. Demonstrate an understanding of the possible role consolidation versus observation after Initial Therapy Target Audience This activity targets physicians in the fields of oncology and hematology. Accreditation This activity has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education through the joint sponsorship of CME LLC and The Oncology Group. CME LLC is accredited by the ACCME to provide continuing medical education for physicians. Continuing Education CreditAMA PRA Category 1 Credit™ CME LLC designates this educational activity for a maximum of 2 AMA PRA Category 1 Credits™ . Physicians should only claim credit commensurate with the extent of their participation in the activity. Compliance Statement This activity is an independent educational activity under the direction of CME LLC. The activity was planned and implemented in accordance with the Essential Areas and policies of the ACCME, the Ethical Opinions/Guidelines of the AMA, the FDA, the OIG, and the PhRMA Code on Interactions with Healthcare Professionals, thus assuring the highest degree of independence, fair balance, scientific rigor, and objectivity. However, CME LLC, the Grantor, and CMPMedica shall in no way be liable for the currency of information or for any errors, omissions, or inaccuracies in the activity. Discussions concerning drugs, dosages, and procedures may reflect the clinical experience of the author(s) or may be derived from the professional literature or other sources and may suggest uses that are investigational in nature and not approved labeling or indications. Activity participants are encouraged to refer to primary references or full prescribing information resources. The opinions and recommendations presented herein are those of the author(s) and do not necessarily reflect the views of the provider or producer. Financial Disclosures Dr. Langer has received grant and research support from Bristol-Myers Squibb, ImClone, Pfizer, Lilly, Schering-Plough Research Institute, Sanofi-Aventis, Amgen, Cell Therapeutics Inc, OrthoBiotech, Celgene, Vertex, Genentech, OSI, AstraZeneca, Active Biotech, Medimmune (absorbed by AZ 2008); has served as scientific advisor for Bristol-Myers Squibb, ImClone, Sanofi-Aventis, Pfizer, IntraBiotics, GlaxoSmithKline, Pharmacyclics, Amgen, AstraZeneca, Novartis, Genentech, Savient, Bayer, Onyx, Abraxis, and Abbott; and has served on the speaker’s bureau for Bristol-Myers Squibb, Sanofi-Aventis, Lilly, OrthoBiotech, Genentech, and OSI. Dr. Simon has no financial relationships to disclose. Copyright Copyright owned by CME LLC. Copyright 2008, all rights reserved. Contact Information We would like to hear your comments regarding this or other activities provided by CME LLC. In addition, suggestions for future activities are welcome. Contact us at: Director of Continuing Education CME LLC Harborside Financial Center Plaza 3, Suite #806 Jersey City, NJ 07311 Dear Colleague: As you well know, our empiric therapy of non-small cell lung cancer (NSCLC) is inadequate. Median survival in advanced disease with standard chemotherapy remains mired at 10 to 12 months, and fewer than 15% to 20% live beyond 2 years. In locally advanced NSCLC, median survival in the setting of combined chemotherapy and radiation has improved to 18 to 24 months, but in the absence of surgery, less than one in five patients will enjoy long-term survival. Even in resectable disease, less than one half of patients who undergo resection are cured. Under these circumstances, individualized or customized therapy employing proteomic and molecular markers may hold the key to improving outcome in this challenging disease. In our continuing series of E-Updates on the controversies and challenges in the treatment of NSCLC, we are pleased to present a robust discussion on the role of new tissue markers in the management of NSCLC by Dr. George Simon, an acknowledged expert in this area. His E-Update is entitled “Molecular Profiling for Cytotoxics and Targeted Agents in NSCLC: Ready for Prime Time?” Several molecules and profiles are emerging with promising utility as predictive and prognostic parameters in NSCLC independent of the standard clinical parameters, such as stage, performance status, and gender. Most notably, these include the genes ERCC1 and RRM1 which are involved in nucleotide metabolism and DNA damage repair, EGFR which is involved in cell proliferation and survival, and oligonucleotide expression array profiles which are signatures of global gene expression associated with specific tumor phenotypes. In addition, proteomics using MALDI and gene expression profiles may prove both prognostic and predictive and are now undergoing research as tools to optimize therapy. Dr. Simon explores the background of these markers, clinical-pathologic correlative work to date, and potential practical applications in the setting of NSCLC. I am sure you will find this review both concise and useful. This last issue of these lung cancer E-Updates discussed the current controversies regarding the role of combined modality therapy in the treatment of locally advanced, unresectable NSCLC. I hope you will take time to read these reviews and provide us the all important feedback. As always, please feel free to contact me with your comments and questions at: corey.langer@uphs.upenn.edu. Hope all of you are having a good and enjoyable holiday season. Best regards, Corey J. Langer MD, FACP Professor of Medicine Director of Thoracic Oncology University of Pennsylvania Philadelphia, Pennsylvania Summary Lung cancer is the leading cause of cancer-related mortality. Improved understanding in the molecular biology and genetics of lung cancer has resulted in the identification of individual genes, gene expression profiles, and molecular pathways that may be useful for clinical management decisions. This E-Update focuses on recent molecules and platforms under evaluation for implementation for use in clinical decision making. Prognostic molecular parameters are defined as markers that affect overall outcome in terms of survival, independent of therapeutic interventions. Predictive molecular parameters are defined as markers that influence therapeutic efficacy. Several molecules and profiles are emerging with promising utility as predictive and prognostic parameters in non-small-cell lung cancer (NSCLC) independent of the standard clinical parameters, such as stage, performance status, and gender. Most notably, these include the genes ERCC1 and RRM1, which are involved in nucleotide metabolism and DNA damage repair; EGFR, which is involved in cell proliferation and survival; and oligonucleotide expression array profiles, which are signatures of global gene expression associated with specific tumor phenotypes. Introduction Lung cancer is the leading cause of cancer-related mortality worldwide. Approximately 85% of lung cancer cases are non-small-cell type (NSCLC). Surgery, chemotherapy, and radiotherapy have been used in various combinations to improve survival and to maximize the therapeutic benefit. Despite advances in treatment, the 5-year survival rate for NSCLC across all stages is currently only 16%. Gene mutations triggered by environmental exposure and intrinsic genome plasticity are thought to be responsible for cancer development. Recent advances in the identification of molecular determinants of poor outcome (prognostic) and therapeutic benefit (predictive) for patients with NSCLC hold the promise for rational clinical decisions based on individual patients’ molecular tumor profiles. The terms “prognostic” and “predictive” are often used interchangeably. However, the term “prognostic” specifically refers to a marker/parameter that is useful for estimating patients’ prognosis (outcome) independent of therapeutic decisions; i.e., survival is different in marker positive and negative patients regardless of intervention. The term “predictive” refers to a marker/parameter that is useful in making therapeutic decisions, i.e., the effect of treatment is different in marker positive and negative patients. For instance, female gender is both prognostic and predictive of outcome. .Women tend to have a better survival than men independent of therapy. Female gender is also predictive of improved response to EGFR tyrosine kinase inhibitors; women have a higher likelihood of response to these agents compared to men. Similarly, elevated ERCC1 (excision repair cross complementation) levels in tissue are both prognostic and predictive. In the absence of therapy, elevated levels are associated with better survival, but they are also predictive of reduced response to platinum-based therapy. Prognostic Markers in Patients with NSCLC: ERCC1 and RRM1 as Markers of Improved Prognosis in NSCLC ERCC1 is a DNA damage repair gene that encodes the 5’ endonuclease of the NER (nucleotide excision repair) complex.[1] Cisplatin causes cytotoxicity of cancer cells by forming adducts that result in DNA cross-links. The NER complex recognizes and removes these adducts, and may thus trigger resistance to platinum agents (Figure 1).
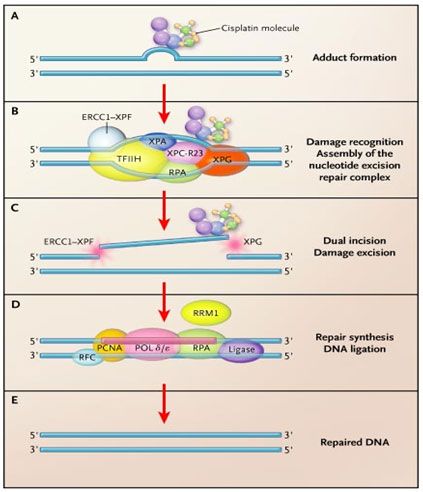
Figure 1: ERCC1 and RRM1 in DNA damage repair. Panel A: Cisplatin binds covalently to genomic DNA, forming a bulky, helix-distorting adduct. The most prevalent adduct is the intrastrand linkage of two adjacent guanine bases by the nitrogen atoms at position 7 (the GG adduct). In chemosensitive cells with low nucleotide excision repair activity, apoptosis usually follows. In chemoresistant cells with high nucleotide excision repair activity, the adduct may be excised and the DNA repaired. Panel B: The adduct is recognized, and proteins of the nucleotide excision repair complex are assembled at the adduct site. The heterodimeric protein excision repair cross-complementation group 1 (ERCC1)–XPF is the last component to be assembled. It is the rate-limiting step. Unwinding of the DNA duplex in the immediate vicinity of the adduct results in the formation of a bubble. Panel C: Endonucleases create dual incisions flanking the damaged bases, with the protein XPG acting on the 3' side and the heterodimer ERCC1–XPF acting on the 5' side. A segment of about 22 to 32 nucleotides containing the adduct is removed. Panel D: The excised segment is repaired by polymerases and the accessory replication proteins PCNA, RPA, and RFC. The integrity of the damaged strand is restored by DNA ligase. Panel E: The repair process is complete, and the original state of the DNA is restored. Ribonucleotide reductase, although not an integral part of the repair complex, catalyzes the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides, providing the building blocks for reconstitution of the excised oligonucleotide. (Modified from N Engl J Med 356: 771-772, 2007; editorial to accompany.[3])
Investigators at Moffitt Cancer Center evaluated the effect of intratumoral ERCC1 expression on survival in 51 NSCLC patients who underwent surgical resection for cure.[2] ERCC1 mRNA expression was a continuous parameter, and the value of 50 (cohort median) was used to dichotomize patients into high and low ERCC1 expressors. A statistically significant difference (p=.01) in median survival was seen in patients with high ERCC1 expression (94.6 months) compared to patients with low ERCC1 expression (35.5 months). A significant relationship was also observed between ERCC1 expression and survival when the levels of ERCC1 were categorized into <30, 30-100, and >100. The median survival in these groups was 35.5, 62.1, and 94.6 months, respectively (Figure 2).
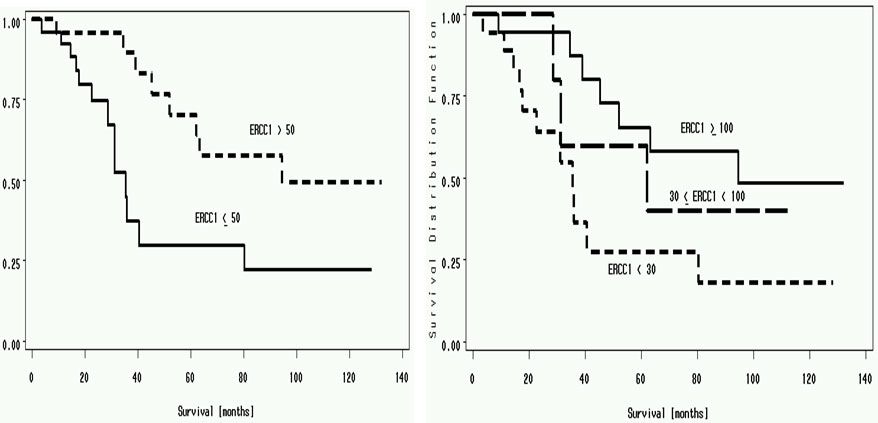
Figure 2:Panel A: Median survival of patients with ERCC1 of more than 50 (94.6 months) vs. less than 50 (35.5 months) (p=.01). Panel B: Median survival of patients with ERCC1 of less than 30 (35.5 months), 30 to 100 (62.1 months) and >100 (94.6 months) (p=.03).[2]
These findings were confirmed by Olaussen et al [3] in a cohort of specimens from the International Adjuvant Cancer Trial (IALT). In this study immunohistochemical (IHC) staining was used to determine the expression of ERCC1 protein in 761 resected NSCLC tumors. These patients had been randomized to treatment with cisplatin-based chemotherapy or observation. Patients were dichotomized into high and low ERCC1 expressors by using the median level as cut-off. Results of this study validated the mRNA expression-based findings of our previous study. Patients in the control group, who had not received adjuvant chemotherapy with ERCC1 positive tumors, had longer survival compared to patients with ERCC1 negative tumors (adjusted HR 0.66; 95% confidence interval [CI], 0.49 to 0.90; p=.009). Among patients who received adjuvant chemotherapy, a significantly prolonged survival was observed in ERCC1-negative tumors (adjusted HR, 0.65; 95% CI, 0.50 to 0.86; p=.002), but not among patients with ERCC1 positive tumors (adjusted HR 1.14; 95% CI, 0.84 to 1.55; p=.40) (Figure 3).
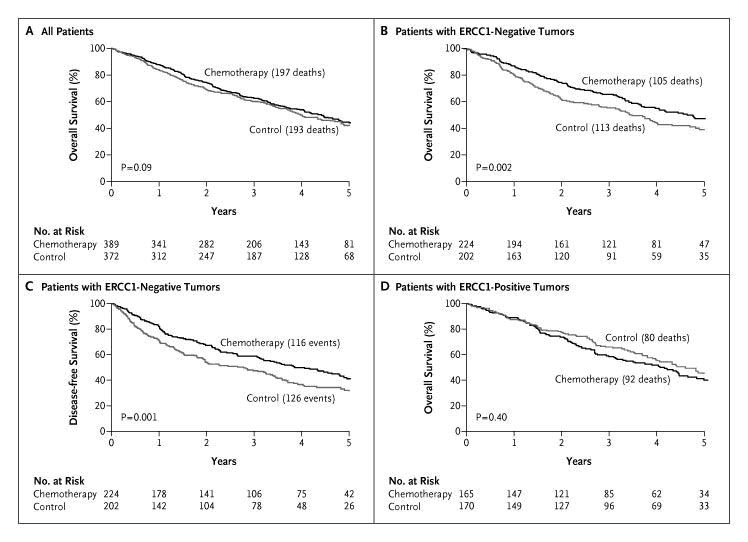
Figure 3: Overall survival of patients analyzed for ERCC1 expression from the International Adjuvant Lung Trial.[3] Panel A: Overall survival of all 761 patients for whom ERCC1 expression data based on IHC analysis were obtained by assigned treatment. Panel B: Overall survival was significantly better in patients with low ERCC1 expression who received adjuvant chemotherapy versus observation (p=.002). Panel C: Disease-free survival was also significantly better in patients with low ERCC1 expression who received chemotherapy versus observation (p=.001). Panel D: Patients with high ERCC1 expression did not benefit from cisplatin-based chemotherapy compared to observation (p=.40).
The study concluded that low ERCC1 expression was associated with a survival benefit from cisplatin-based adjuvant chemotherapy for resected NSCLC. Patients with high ERCC1 in the control group had a better outcome than those with low ERCC1; yet their tumors were resistant to platinum-based therapy. These observations identified a group of patients (those with low ERCC1) that were likely to derive maximal benefit from platinum-based adjuvant chemotherapy. Another enzyme involved in DNA synthesis and repair is ribonucleotide reductase, which catalyzes the biosynthesis of deoxyribonucleotides from corresponding ribonucleotides.[4] It is the molecular target of gemcitabine, an antimetabolite with activity in several malignancies including NSCLC. RRM1, the regulatory subunit of ribonucleotide reductase, plays a role in suppressing tumor cell migration and metastasis formation, probably through the tumor suppression gene PTEN, which is involved in the attenuation of growth factor signaling through its lipid and protein phosphatase activity.[5] Decreased survival has been observed in NSCLC patients with loss of one copy of the RRM1 gene compared to those with two intact copies [6], and similar prognostic function has been demonstrated with RRM1 as with ERCC1. Oligonucleotide-based Gene Expression Signatures of Improved Survival in Resected NSCLC Patients With the advent of whole genomic and/or proteomic approaches to classify cancers, the development of molecular profiles as prognostic markers of outcome or predictive markers of response to therapy has increased substantially. Oligonucleotide array-based gene expression patterns have been studied to evaluate and develop the use of the gene expression profiles as a means to stratify risk and treatment.[7] In one study from a cohort of 89 patients with early-stage NSCLC, a profile was developed and validated independently in two groups of patients: 25 patients from the American College of Surgeons Oncology Group (ACOSOG) Z0030 study and 84 patients from the Cancer and Leukemia Group B (CALGB) 9761 study. The authors concluded that their expression profile was prognostic of recurrence for individual patients with an accuracy of 72% and 79% in the two groups, respectively. Prognostically, it was a better marker of 5-year survival than clinical or pathological stage. Thus, the profile may provide a method to identify patients at high risk of recurrence who may benefit from adjuvant chemotherapy. Chen et al. [8] developed a similar signature that was associated with survival in 125 randomly selected patients. By using oligonucleotide microarray data and risk scores, 16 genes were identified that correlated with survival. Of these 16 genes, five genes (DUSP6, MMD, STAT1, ERBB3, and LCK) were selected for reverse-transcriptase polymerase chain reaction (RT-PCR) and decision tree analysis; together, they were independent predictors of progression-free and overall survival. A significant correlation was observed between the microarray-based and RT-PCR-based expression levels for the five genes in 101 patients. The use of the authors’ profiles resulted in separating patients into high risk (59 patients) and low risk (42 patients) groups. The five-gene signature was strongly associated with overall survival with 96% accuracy, 98% sensitivity, 93% specificity, and 98% positive predictive value. Patients with a high-risk gene signature had a shorter overall survival compared to patients with a low-risk signature (20 months vs. 40 months, p<.001) (Figure 4).
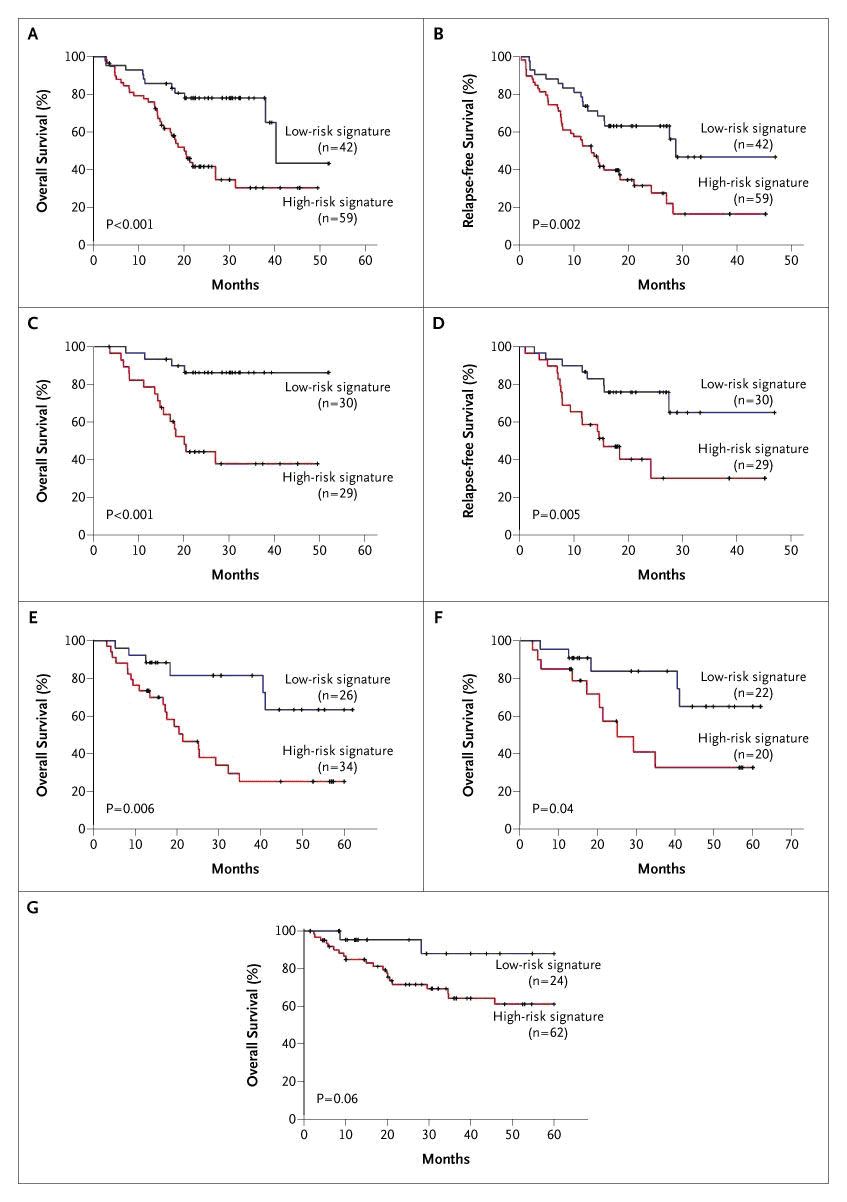
Progression-free survival in the high-risk group was 13 months, whereas the low-risk group had a median progression-free survival of 29 months (p=.002). Serum Proteomic Profiles as Prognostic Marker of Improved Prognosis in NSCLC Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-MS) holds the promise to produce high-quality, cancer-specific protein profiles.[9] Proteomic patterns obtained directly from small amounts of fresh frozen lung-tumor tissues have been used to study associations with tumor type, nodal involvement, and survival of cancer patients. In a study by Yanagisawa et al [10], proteomic spectra were obtained from 79 lung tumors and 14 normal lung tissues to build and test a class-prediction profile. Profiles based on differentially expressed peaks were able to classify lung cancers by histology, distinguish primary lung cancers from cancers metastatic to the lung, and determine nodal involvement with 85% accuracy. Based on a profile of 15 distinct mass spectrometry peaks, patients were differentiated into those with poor prognosis (median survival 6 months, N=25) and those with good prognosis (median survival 33 months, N=41, p<.0001). Markers That Predict for Therapeutic Efficacy The current standard of treatment for patients with incurable metastatic NSCLC is a doublet chemotherapy regimen.[11] The principal agents used are cisplatin or carboplatin, docetaxel or paclitaxel, gemcitabine, and vinorelbine. The reported response rates to such therapy are 17%–37%, the median overall survival ranges from 6.7–11.3 months, and the 1-year and 2-year survival rates are 31%–46% and 9%–21%, respectively. When tumors progress during or after initial therapy, response to further systemic treatment is approximately 10% for single agent therapy.[12] Variable response and survival in this setting suggests that resistance to systemic therapy is probably not an all-or-none phenomenon, but rather a function of the molecular characteristics of the individual tumor. ERCCI as a Predictor for Platinum Resistance in Advanced NSCLCPlatinum compounds are heavy metal complexes that form adducts with and cross-links between DNA molecules and thus effectively block DNA replication and transcription. Repair of these adducts and cross-links are dependent on ERCC1. In gastric cancer, ERCC1 mRNA levels are inversely associated with response and survival to platinum-containing treatment.[13] The median survival in patients with low ERCC1 was estimated to be more than 24 months. On the other hand, patients with high ERCC1 expression had a median survival of 5.4 months (p=.034). Similar observations have been reported for malignancies of the ovaries [14], esophagus, colorectum [15], lung [16, 17] and breast [18]. In these studies, ERCC1 levels were evaluated by RT-PCR, on gel-based or real-time-based platforms, using specimens that were preserved in formalin or by freezing. Lord et al. [16], correlated response and survival with the level of ERCC1 expression in 56 patients with advanced NSCLC treated with gemcitabine and cisplatin. mRNA was isolated from formalin-fixed tumor specimens prior to therapy, and relative expression levels of ERCC1 were determined by real-time RT-PCR. The overall response rate was 44.7%, and there was no significant correlation with ERCC1 levels. Median overall survival was significantly longer in patients with low ERCC1 expression (61.6 weeks; 95% CI, 42.4–80.7) compared to patients with high expression (20.4 weeks, 95% CI, 6.9–33.9 weeks), (p=.046). As a result, it is unclear if there was an interaction between ERCC1 expression and treatment, i.e., the predictive utility of ERCC1. As mentioned earlier, Olaussen et al. found that ERCC1 protein expression was associated with benefit from adjuvant cisplatin-based therapy (p=.009) in a large group of patients with surgically resected NSCLC. ERCC1 was predictive of the benefit of adjuvant cisplatin-based chemotherapy. Only patients with low tumoral ERCC1 protein levels benefited from adjuvant chemotherapy (adjusted hazard ratio for death, 0.65; 95% CI, 0.50 to 0.86; p=.002). Investigators at Moffitt had studied prospectively if an association between ERCC1 mRNA levels as determined by real-time RT-PCR in fresh-frozen tumor specimens and response to two cycles of gemcitabine/carboplatin exists. In a group of 35 patients, we found an inverse correlation between ERCC1 levels and tumor shrinkage (r = -0.283, p=.099).[19] RRMI as a Predictor of Gemcitabine Efficacy Several studies have demonstrated that RRM1 is a molecular target of gemcitabine and thus a key cellular determinant of its therapeutic efficacy. To evaluate the impact of intratumoral RRM1 expression on the efficacy of gemcitabine and carboplatin in previously untreated NSCLC patients, Moffitt investigators conducted a prospective phase II clinical trial [19], to confirm these in vitro data. Findings of this study confirmed that tumoral levels of RRM1 expression were significantly and inversely correlated with the magnitude of tumor shrinkage (r = -0.498, p=.002). BRCA1 as a Predictor of Chemotherapy Efficacy BRCA1 plays an important role in DNA repair pathways. It functions as a sensitizer to apoptosis induced by antimicrotubulin agents such as taxanes and vinca alkaloids and conversely mediates resistance to DNA-damaging agents such as platinum compounds. Taron et al. reported the first clinical study on the potential predictive value of BRCA1 mRNA levels in NSCLC patients treated with gemcitabine and cisplatin.[20] BRCA1 levels were detected in all 55 resected tumors, and they ranged from 0.28 to 10.43. Patients were grouped based on the BRCA1 expression (<0.61, .65-2.37, >2.45). Median survival was longer in the low expression group compared to the high expression group (not reached in the low group, 12.7 months in the high group with a 95% CI of 0.28-28.8 months; p=.01). Because of the study design, the impact of BRCA1 expression on treatment efficacy could not be assessed in detail. As a result, the predictive value of BRCA1 levels on chemotherapeutic efficacy remains unclear. Predictors of Improved Response or Survival with EGFR Tyrosine Kinase Inhibitors The epidermal growth factor receptor (EGFR) protein is a member of the HER (referred to as “erb-B” in mouse studies) family of receptor tyrosine kinases that are involved in the pathogenesis of many cancers, including NSCLC.[21] The orally active, selective EGFR tyrosine kinase inhibitors (EGFR-TKI) gefitinib and erlotinib have yielded response rates of 9% to 26% in advanced stage NSCLC. Lynch et al. and Paez et al. reported mis-sense mutations and deletions in the tyrosine kinase domain of EGFR gene which were highly associated with gefitinib response.[22, 23] These somatic mutations were small, in-frame deletions, or amino acid substitutions clustered around the ATP-binding pocket of the tyrosine kinase domain, and they were predominantly identified in patients with gefitinib-responsive tumors. These data, and other subsequently reported studies, demonstrate the predictive utility of EFGR mutations for EGFR-TKIs. Hirsch et al. [21] assessed EGFR and HER2 gene copy numbers by fluorescence in situ hybridization (FISH) in 81 patients with advanced bronchioloalveolar carcinoma treated with gefitinib. Tumors were dichotomized into EGFR FISH-positive and negative groups. Sixty-three percent of FISH-positive patients had non-progressive disease as best response compared to 39% of FISH-negative patients (p=.087). Improved survival was also observed in patients with increased EGFR gene copy numbers. This analysis strongly suggested the predictive utility of EGFR gene copy numbers for gefitinib efficacy. Oligonucleotide-based Gene Expression Profiles as Predictors of Response in NSCLC Patients Using in vitro drug sensitivity and oligonucleotide expression data, Potti et al. [24] developed gene signatures to predict sensitivity to individual chemotherapeutic drugs. Independent sets of cell lines were identified, and each signature was validated with response data. A gene expression-based predictor was developed consisting of 50 genes that classified cell lines based on docetaxel sensitivity. The docetaxel signature predicted sensitivity with an accuracy of 80% in an independent dataset (p<.001). The study also integrated chemotherapy response signatures with oncogenic pathway deregulation to obtain information on potential drug efficacy in the context of specific pathways involved in tumorigenesis. Oligonucleotide expression array data from 17 NSCLC cell lines predicted to be resistant to docetaxel also had profiles suggestive of phosphotidylinositol 3 kinase (PI3K) pathway activation. Cell lines with the PI3K activation profile responded to PI3K inhibitors. Individualizing Treatment for Advanced Stage NSCLC Using ERCC1 and RRM1 We recently reported results from a prospective phase II clinical trial that was designed to test the feasibility and efficacy of molecular analysis-directed individualized therapy in patients with advanced NSCLC.[25] ERCC1 and RRM1mRNA expression levels were determined by real-time RT-PCR in fresh-frozen tumor specimens from 53 eligible patients. Predetermined values for ERCC1and RRM1 were used to select or omit the drugs gemcitabine and carboplatin in the treatment of advanced, chemotherapy-naive NSCLC patients. Gemcitabine was used in the treatment doublet if RRM1 was ≤16.5, and carboplatin was used in the doublet if ERCC1 was ≤ 8.7. This strategy resulted in four gene expression groups. The low RRM1 and low ERCC1 group was treated with gemcitabine and carboplatin (GC group); the low RRM1 and high ERCC1 group was treated with gemcitabine and docetaxel (GD group); the high RRM1 and low ERCC1 group was treated with docetaxel and carboplatin (DC group); and the high RRM1 and high ERCC1 group was treated with vinorelbine and docetaxel (DV group). Disease response rate was 42%. Overall and progression-free survivals were 59% and 14% at 12 months with medians of 13.3 and 6.6 months, respectively. Overall survival did not vary with the assigned chemotherapy (Figure 5).
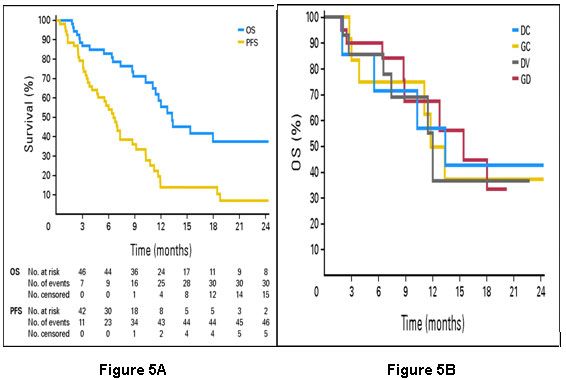
Figure 5:Panel A: Overall survival (OS) and progression-free survival (PFS) of 53 patients with advanced non–small-cell lung cancer treated with chemotherapy based on expression of the genes ribonucleotide reductase subunit 1 (RRM1) and excision repair cross-complementing group 1 gene (ERCC1). Panel B: Overall survival (OS) by assigned chemotherapy. DC, docetaxel and carboplatin; GC, gemcitabine and carboplatin; DV, docetaxel and vinorelbine; GD, gemcitabine and docetaxel. Adapted with permission from Simon et al.[25]
The conclusion from this trial was that gene expression analysis for treatment decisions of individual patients with advanced NSCLC was feasible, that regimens selected in this manner were safe, and that it produced favorable response and survival data. These promising findings require confirmation in a randomized phase III trial. Such a confirmatory phase III trial is currently underway. Cobo et al. [26] recently reported results from the first customized and randomized trial in patients with advanced NSCLC. In this trial, ERCC1 mRNA expression, determined by real-time RT-PCR in routinely collected diagnostic tumor specimens, was used to decide whether cisplatin was included in a doublet regimen. The primary study endpoint was best disease response. Of 366 patients that had a successful gene expression analysis and received at least one cycle of therapy, 346 patients were evaluable for response. The response rate was 39.3% in the control arm (docetaxel and carboplatin), while it was 50.7% in the experimental arm (docetaxel and gemcitabine if ERCC1 was high; docetaxel and carboplatin if ERCC1 was low). However, there were no differences in overall survival between the two groups. Conclusions The level of ERCC1 expression is prognostic of survival and a predictive marker for platinum efficacy. Elevated levels of ERCC1 are associated with longer survival; however, they are also predictive of lack of platinum efficacy. Likewise, the level of RRM1 expression is prognostic of survival and a predictive marker for gemcitabine efficacy. BRCA1 expression levels are a potentially important tool for the selection of chemotherapeutic agents. EGFR mutations and gene copy numbers are prognostic of survival and predictive of EGFR-TKI efficacy. The presence of mutations and high gene copy numbers are also predictive of EGFR-TKI efficacy. Oligonucleotide expression array and proteomic profiles are promising alternative strategies for outcomes prognostication and prediction of therapeutic efficacy. All of these approaches require further development before we can safely adopt a customized or individualized approach to the management of patients with advanced NSCLC.
References
1. de Laat WL, Jaspers NG, Hoeijmakers JH: Molecular mechanism of nucleotide excision repair. Genes Dev 13:768-785, 1999. 2. Simon GR, Sharma S, Cantor A, et al: ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 127:978-983, 2005. 3. Olaussen KA, Dunant A, Fouret P, et al: DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355:983-391, 2006. 4. Elledge SJ, Zhou Z, Allen JB: Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci 17:119-123, 1992. 5. Gautam A, Li ZR, Bepler G: RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene 22:2135-2142, 2003. 6. Bepler G, Sharma S, Cantor A, et al: RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol 22:1878-1885, 2004. 7. Potti A, Mukherjee S, Petersen R, et al: A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 355:570-580, 2006. 8. Chen HY, Yu SL, Chen CH, et al: A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356:11-20, 2007. 9. Amann JM, Chaurand P, Gonzalez A, et al: Selective profiling of proteins in lung cancer cells from fine-needle aspirates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Cancer Res 12:5142-5150, 2006. 10. Yanagisawa K, Shyr Y, Xu BJ, et al: Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362:433-439, 2003. 11. Schiller JH, Harrington D, Belani CP, et al: Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346:92-98, 2002. 12. Hanna N, Shepherd F, Fossella FV, et al: Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J Clin Oncol 22:1589-1597, 2004. 13. Metzger R, Leichman CG, Danenberg KD, et al: ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol 16:309-316, 1998. 14. Dabholkar M, Bostick-Bruton F, Weber C, et al: ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl CancerInst 84:1512-1517, 1992. 15. Shirota Y, Stoehlmacher J, Brabender J, et al: ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol 19:4298-4304, 2001. 16. Lord RV, Brabender J, Gandara D, et al: Low ERCC1 Expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 8:2286-2291, 2002. 17. Lord RVN, Brabender J, Gandara D, et al: Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 8:2286-2291, 2002. 18. Bewick MA, Conlon MS, Lafrenie RM: Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol 24:5645-5651, 2006. 19. Bepler G, Kusmartseva I, Sharma S, et al: RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol 24:4731-4737, 2006. 20. Taron M, Rosell R, Felip E, et al: BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 13:2443-2449, 2004. 21. Hirsch FR, Varella-Garcia M, McCoy J, et al: Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol 23:6838-6845, 2005. 22. Paez JG, Janne PA, Lee JC, et al: EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304:1497-1500, 2004. 23. Lynch TJ, Bell DW, Sordella R, et al: Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350:2129-2139, 2004. 24. Potti A, Dressman HK, Bild A, et al: Genomic signatures to guide the use of chemotherapeutics. Nat Med 12:1294-1300, 2006. 25. Simon G, Sharma A, Li X, et al: Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. Journal of Clinical Oncology 25:2741-2746, 2007. 26. Cobo M, Isla D, Massuti B, et al: Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol 25:2747-2754, 2007.
