Advances in a wide array of scientific technologies have brought data of unprecedented volume and complexity into the oncology research space. These novel big data resources are applied across a variety of contexts-from health services research using data from insurance claims, cancer registries, and electronic health records, to deeper and broader genomic characterizations of disease. Several forms of big data show promise for improving our understanding of racial disparities in breast cancer, and for powering more intelligent and far-reaching interventions to close the racial gap in breast cancer survival. In this article we introduce several major types of big data used in breast cancer disparities research, highlight important findings to date, and discuss how big data may transform breast cancer disparities research in ways that lead to meaningful, lifesaving changes in breast cancer screening and treatment. We also discuss key challenges that may hinder progress in using big data for cancer disparities research and quality improvement.
Introduction
For more than 4 decades, a survival gap has persisted between black and white women with breast cancer in the United States; age-adjusted mortality rates are 28 per 100,000 among black women and 20 per 100,000 among white women.[1] Much of what we know about the root causes of this disparity, as well as possible solutions, comes from research using different types of data, ranging from high-dimensional genomic data to large population-based data sources and linkages with insurance claims records. We will provide a brief orientation on the research that has defined our understanding of breast cancer disparities to date, as well as promising emerging data sources and methods that may take us further in the quest to close the racial survival gap and provide better cancer care to vulnerable populations.
What Is the Meaning of Big Data for Oncology Research?
The term "big data" is used in many different industries. It is colloquial rather than scientific, with varying meaning based on the context in which the term is used. A general definition of big data centers on three defining features: size, complexity, and technology.[2] That is, the data are of such enormous quantity and complexity that “manipulation and management present significant logistical challenges” using traditional data computing and technology methods. Big data are also frequently described using 4 to 10 key attributes[3] known as the “Vs” of big data. The four most common Vs are volume, velocity, variety, and veracity.[4] Depending on the field of research, each of these “V” attributes carries a different level of importance, and presents different challenges, to data users. Extensive research and technology developments are now emerging that help data users deal with each of the big Vs in their efforts to transform large and complex datasets into valuable and actionable information.
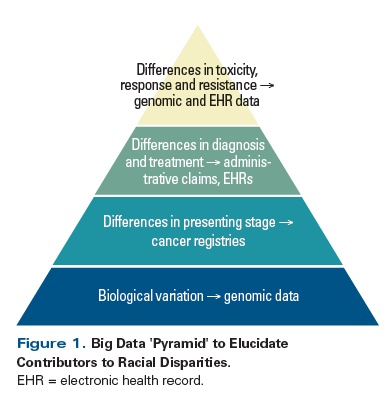
In cancer disparities research, big data resources can be powerful because the size (volume) and breadth (variety) of data captured means that, compared with clinical trial data or data from academic centers or consortia, the information may better represent vulnerable patient groups, such as minorities or elderly patients. Similarly, the tools and analytic methods developed for working with these data can model and measure how multiple complex factors interact to reveal specific disparities in care. For the purposes of this discussion, we will highlight the use of certain big data resources and analytic methods for cancer disparities research, including large linked administrative and cancer registry datasets, aggregated data from electronic health records (EHRs), and genomic data. A model for integrating these data sources to fully understand cancer disparities is illustrated in Figure 1.
Major Contributors to Racial Disparities in Breast Cancer
It is widely acknowledged that both social and biological factors contribute to the survival gap between black women and white women with breast cancer.[5] The epidemiologic basis of racial disparities in breast cancer has been defined using data from the Surveillance, Epidemiology, and End Results (SEER) Program national network of cancer registries, as well as analyses of national cancer incidence and mortality data from the American Cancer Society.[1] We know from these large, longitudinal, national data sources that black women, particularly those under 50 years of age, have a disproportionately large burden (measured as relative frequency) of the biologically aggressive triple-negative breast cancer subtype, as well as a more advanced disease stage at presentation.[6,7] Genomic sequencing data, which enable more precise molecular characterization of breast cancers, have been used in studies such as the Carolina Breast Cancer Study and The Cancer Genome Atlas project, to identify additional biological differences within clinically defined groups, including higher proportions of poor-prognosis molecular subtypes among young black women whose clinical markers indicate hormone receptor (HR)-positive and human epidermal growth factor receptor 2 (HER2)-negative disease. This variation within clinical subtypes may contribute to racial disparities in breast cancer outcome in these HR-positive HER2-negative patients, who would usually be expected to have a favorable prognosis.[8-10]
While unrecognized biological variation likely explains a portion of within-subtype variability in survival outcomes, we also have ample documentation, primarily from large cancer registry datasets linked to administrative claims data, that treatment disparities are prominent in breast cancer care and contribute to differences in disease outcomes. Beginning with surgical therapy, black women are less likely to receive any definitive surgery for early-stage disease,[11] and less likely to receive morbidity-sparing sentinel lymph node biopsy when eligible.[12] There is substantial evidence that these disparities are partially explained by the concentration of surgical treatment for black patients within lower-volume and lower-quality hospitals that are less likely to be integrated into research networks.[13-15] Adjuvant breast radiation therapy (RT) is more often delayed among black women; delays are linked to both site of care (with access to smaller surgical facilities and/or those with onsite RT available linked to higher odds of accessing RT, compared with access to large governmental facilities only)[16] and breast cancer mortality.[17] With respect to adjuvant chemotherapy, initiation of chemotherapy appears to be relatively equal by race. However, black women more often have delays between surgery and chemotherapy,[18] and may discontinue chemotherapy[19] and biologic therapy[20] prematurely; both treatment patterns are demonstrably related to survival decrements.[18-20] Black women with HER2-overexpressing breast cancer receive trastuzumab, a highly effective but costly targeted therapy, at dramatically lower rates than white women.[21] Among eligible patients, black women initiate adjuvant hormonal therapy at lower rates than white women and have more problems with adherence; this disparity appears to be concentrated among younger patients and patients treated with chemotherapy.[22] With regard to genomic testing, black women are less likely to receive guideline-concordant gene expression profile testing to help predict the benefit from chemotherapy in those with HR-positive disease.[23,24]
Leveraging Healthcare Access Data to Understand Cancer Disparities
Several main categories of big data resources may be useful for studying and improving breast cancer care. The first type is administrative claims data. Generally, these data are obtained for research from large insurance providers (such as the Centers for Medicare and Medicaid Services [CMS], for Medicare and Medicaid data), linked data resources from a state or geographic region (such as the North Carolina Integrated Cancer Information and Surveillance System [ICISS],[25] for claims from multiple payers in the state of North Carolina), or pools of commercial insurance payers who have agreed to aggregate de-identified data (such as the MarketScan database of Truven Health Analytics, an IBM Watson Health company).[26] Strengths of administrative claims data include the detailed and temporal record of treatments provided to patients; and the representation of diverse patients across geography, age, socioeconomic status, and other categories. Claims data follow patients from one site of care to another; these records remain intact despite changes in healthcare providers over time, but there may be disruptions in access if a patient changes insurance providers. Claims generally contain information about costs of care, making them particularly valuable in cost and value analyses. Claims data also generally contain information on both cancer patients and otherwise similar patients without a cancer diagnosis, facilitating the design of case-control studies. Limitations of claims data for cancer disparities research are often the result of a lack of clinical detail regarding cancer stage and other clinical characteristics, as well as outcomes of treatment such as recurrence and survival.
Some limitations of claims data can be obviated by linking with other data sources. A wide variety of other data types have been linked to administrative claims. Data linkages that can enrich understanding of the patient’s disease include cancer registry data, as in the SEER-Medicare and ICISS datasets, patient-reported data such as the Medicare Health Outcomes Survey of beneficiaries,[27] and laboratory data. Linkages of claims and registry data to other sources such as census data and the National Death Index have also enhanced the ability to characterize patients’ socioeconomic situations and their survival after cancer treatment. However, data sources are not available to provide additional information on all desired variables regarding treatment, comorbidities, or other variables likely to affect outcome, and even when linked there may not be adequate patient overlap between the clinical and administrative data. In these cases, researchers can sometimes apply claims-based algorithms to define important exposures, such as the receipt of a certain treatment or the burden of other illnesses, or to define outcomes of interest, such as treatment toxicities, cancer recurrence, or cancer-related death. Depending on the complexity of the algorithm, the availability of a dataset with clinical annotation to validate the algorithm, and the specificity of the billing codes used in the clinical situation of interest, such algorithms can be of greater or lesser value for research. Breast cancer recurrence has been especially challenging to identify accurately in claims, although published and validated algorithms do exist,[28,29] while progression of disease in the metastatic setting is virtually impossible to ascertain. An additional challenge in disparities research is that racial and ethnic minorities are disproportionately uninsured and overrepresented in the databases of public insurers, so that these populations in any one particular claims database may be small and/or not representative of the experience of other patients.
Along with the growing use of EHRs in clinical practice, there is increased leveraging of the vast amounts of data collected within these systems for use in cancer research, including disparities research. Advantages of these data sources include the rich clinical detail not available in claims, the representation of patients across the spectrum of insurance types and age ranges, and the potential to provide accurate information about cancer outcomes. The original research using pooled EHR data was performed in integrated health systems such as Kaiser Permanente and the Group Health Cooperative. Efforts are now underway to aggregate EHR data from oncology practices and hospitals, with a specific focus on providing data for cancer research and quality improvement.
Leveraging Biological Data to Understand Cancer Disparities
A substantial contribution to racial disparities in mortality is differential access to healthcare, which can be directly tracked using the previously discussed resources. However, racial disparities can also arise from differences in tumor biology, and this factor requires distinct types of big data to elucidate. Differences in exposure history over a lifetime, genetic background, and social factors may lead to differences in breast tumor biology or clinical subtype. For example, as discussed, black women have higher relative frequency of all aggressive breast cancer subtypes (luminal B, HER2-enriched, basal-like) and lower relative frequency of the most treatable, indolent luminal A breast cancers.[6,9,10] In the decade since these relative frequency differences were first identified, there has been an evolution in the technology that we use to identify specific tumor subtypes. In early reports of racial disparities by subtype,[6,8] immunohistochemistry data were collected for five markers (estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2, cytokeratin 5/6, and epidermal growth factor receptor), and these immunohistochemistry surrogates were used to approximate genomic subtypes. However, the use of only three to five markers to approximate subtypes that were originally identified across thousands of gene expression features is an error-prone approach that several recent studies have demonstrated can result in substantial misclassification.[30,31] In research settings, initiatives such as The Cancer Genome Atlas project have begun to identify biological subtypes using a broad range of genomic markers, including DNA-sequencing, RNA-sequencing, DNA-methylation, microRNA-sequencing, and protein arrays. Even histologic images of breast tumors can be mined to extract many visual patterns and features, and new prognostic algorithms have been developed through multidimensional analysis of data from such images.[32] In research settings, the number of features that can be used to classify tumors has increased by many orders of magnitude. Different data types can also be brought together in complex ways to identify both commonalities and differences between tumors from different sites.[33]
Some of the most exciting recent advances, and groundbreaking changes in our understanding of cancer, have come from leveraging parts of complex genetic datasets that were previously considered uninformative. For example, previous research has focused on gene-specific “driver” mutational events that lead to specific breast cancer subtypes, and a relatively small number of driver mutations (~30 gene-specific mutations) have been identified, with few racial differences between them. For example, The Cancer Genome Atlas project has elegantly demonstrated that most gene-specific mutations (eg, p53, PIK3CA, etc) are strongly associated with certain clinical subtypes of breast cancer, and that after adjusting for molecular subtype, few differences are detected in mutational frequency by race. More racial differences have been identified when considering gene-independent characteristics, such as the degree of intratumoral heterogeneity, which may be higher in black women.[34] Furthermore, while the number of genes that are frequently mutated across breast cancers is relatively small, the number of mutations harbored in a single cancer genome is manyfold higher, ranging from hundreds to thousands. The biological causes and epidemiology of these mutations are almost entirely unknown, but many of these mutations are simply passenger mutations. Passenger mutations, in contrast to driver mutations, do not have a positive or negative effect on tumor development and are not selected for during development. However, recent research using data from The Cancer Genome Atlas on 30 different cancer types has demonstrated that the patterns of mutations, inclusive of both driver and passenger events, show that there are recurrent mutational signatures that are commonly observed.[35,36] Many of these mutational signatures have been linked to exposure history, and some may eventually be found to be prognostic. Studies have not yet been performed to compare mutational signatures by race. Thus, we have just begun to understand the many ways in which the somatic genomics of black women and white women may progress along different paths to disease. Moving from gene-specific to higher-order relationships among features is challenging and will require continuing methodological advances. Progress in these areas is essential to capturing the full impact of complex genomic data.
Continuing research is generating complex information on the leading edge of data science, and much of this research has already begun to have an impact on clinical practice. Genomic tests such as the Oncotype DX assay, MammaPrint, and the Prosigna Breast Cancer Prognostic Gene Signature Assay have become widely available in clinical settings. Among breast cancer patients with HR-positive, HER2-negative disease, there are a subset with higher risk of recurrence scores, both by Oncotype DX and the Prosigna assays. Wider availability of molecular data would improve our understanding of the heterogeneity of outcomes within a clinically defined group. However, as previously described, barriers to healthcare access have prevented equitable use of such genomic assays in black women. It is likely that advances in genomic science will continue to drive novel, precision medicine approaches for addressing disparities. Leveraging this science will require continued development of data analysis and integration methods, and methods for combining the genomic data with data on healthcare utilization. Data integration has begun to bring together clinical data and genomic data, but extending the technologies employed to include a wider range of data types, over time and across research settings, remains a research frontier.
New Frontiers in Big Data Research: Leveraging Comprehensive Patient Data
Vast amounts of patient data are collected as part of the process of making a diagnosis and administering clinical care. Unfortunately, much of these data exist within discrete and closed information technology systems, and are not easily available to researchers or clinicians seeking to learn from this pooled patient experience. These systems also often contain specific proprietary and non-universal coding nomenclatures or ontologies (eg, CPT [Current Procedural Terminology], LOINC [Logical Observation Identifiers Names and Codes]). For example, information about a given patient’s disease progression or choice of treatment as recoded by clinic notes is captured as unstructured or “free text” data in the EHR and must be restructured and systematically organized for research. Imaging and laboratory information are stored as external images or files that require special software for viewing. Health insurance claims and structured data on diagnosis and procedures are collected from practice and insurance management systems. The mutually exclusive nature of these data systems makes it very difficult to consolidate a complete body of information on an individual patient. The construction of a clinically meaningful “big data” picture of a patient becomes even more complicated as patients move between different providers, clinics, and health systems for diagnoses, treatment, and follow-up care. The field of genomics has benefited from rapid advances in data computing, storage, and analysis but the data remain largely outside the scope of other clinical and computing information systems.
Oncology practices are critical players in the future of big data cancer research. Only with strong participation from both community and academic practices will we be able to fully understand the diversity of disease and related health disparities. Data infrastructure programs such as the American Society of Clinical Oncology’s CancerLinQ initiative[37] and Flatiron Health[38] are able to partner directly with oncology practices and can adapt to different EHR platforms. The NCI Community Oncology Research Program (NCORP) of the National Cancer Institute is also a highly successful federally supported resource that helps to support clinical trial participation in community oncology practice. The predecessor to the NCORP program (the NCI Community Clinical Oncology, or CCOP, program) was effective at helping to diffuse cancer innovation more quickly into community practices and attenuate disparities in the receipt of treatment.[12,14,39,40] In addition to participating in data infrastructure platforms, practicing oncologists are critical stakeholders for patient reporting and disseminating new evidence regarding precision medicine and tumor heterogeneity into real-world practice.
The overarching vision is to create a “learning health system” in which data from real-world cancer patients within the EHR and data from other systems are rapidly analyzed and fed back to the physicians at the point of care.[41] The ultimate goal is to improve the quality of cancer care by using big data and analytics in real time to analyze patient data within a larger context; this approach enables more precise tailoring of treatment and timely delivery of treatment outcomes information to clinicians.
Key Challenges in Applying Big Data to Cancer Disparities Research
Key challenges of using big data resources for cancer disparities research are similar to those complicating use of big data for other goals in cancer care. These challenges can be summarized as follows:
• Data structure and format are not standardized among different systems, significantly complicating the combination of data from multiple sources.
• Information technicians and systems scientists are struggling to develop comprehensive technology platforms that can integrate data across multiple scales and scientific domains (from molecular data to social/behavioral risk factors).
• Large amounts of critical data (eg, cancer stage, recurrence) are trapped in unstructured data fields and require sophisticated human-curation and/or natural language processing approaches to render them usable.
• Gaps exist in data continuity and longitudinal follow-up when portions of patients’ care occur outside of a system, such as a participating provider, clinic, EHR, laboratory, or payer.
While various governmental, academic, and for-profit companies have all initiated big data endeavors, there has been a lack of significant collaboration and data sharing between initiatives. Data use agreements, regulatory measures, and data governance processes, while acting as key safeguards to patient privacy and data security, have hampered efforts at broader sharing of data. Examples of innovations that would ease collaborative sharing of big data in oncology include greater investment by data partners in secure data sharing infrastructures and the use of unified patient identifiers to recognize patients across systems.
KEY POINTS
- Big data that are relevant for oncology disparities research exist in a variety of formats; these include administrative claims, national cancer registries, electronic health records, and genomic data.
- Big data for oncology disparities research are united by their volume, complexity, and requirements for novel technological and analytic methods in order to extract useful and reliable information.
- Analyses based on big data can be compromised when researchers fail to understand the populations, biases, and limitations, as well as the strengths, of each unique data resource. However, analyses can be strengthened and made more representative of vulnerable populations if resources are linked together to offset these limitations.
- The next frontiers of big data research in oncology require increasing collaboration among interdisciplinary teams of clinical, laboratory, and information technology, as well as statistical experts willing to share methods and data between fields in secure, standardized ways.
An additional challenge that is specific to disparities research is the generalizability of patients captured within big data resources. Patients at large centers with sophisticated EHR systems and data-sharing capabilities, even those who belong to minority groups, may differ significantly from minority patients in lower-resource settings, which may not readily share data or leverage electronic resources and/or technology. Likewise, biologic discovery efforts tend to focus on patients with large tumors and ample genomic material for use in multiple genotyping/sequencing platforms. These large tumors, sampled from mostly academic research centers, may not capture the diversity of cases in a broader setting. Thus, study design and possible selection biases should be considered when evaluating and drawing inferences from big data initiatives. It is possible that some of these selection biases shift the frequency of specific tumor types or specific treatment care patterns, but consequences can also be more profound. As the promise of big data is transformed into interventions that improve care quality or close care gaps, disparities could widen if minority patients and minority-serving healthcare providers are not represented in research or infrastructure-building efforts. Special attention and emphasis within big data research are warranted so that information derived from, and specific insights into, minority populations are adequately represented.
The Future: Big Data–Driven Interventions
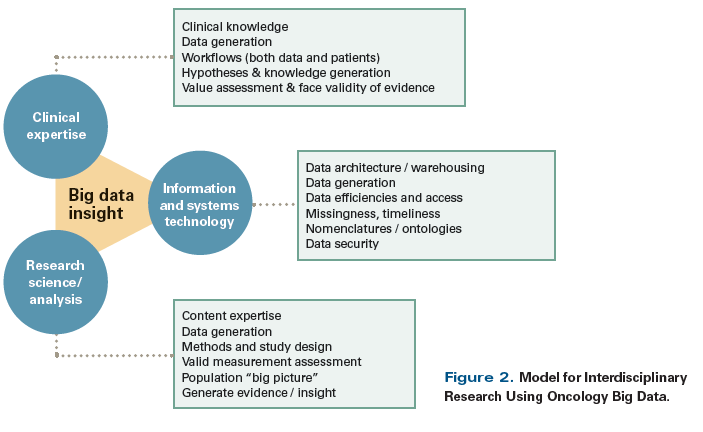
Research in other cancer types has demonstrated that knowledge derived from big data can transform clinical care and/or lead to new interventions. In the translation of complex, high-dimensional genomic data into precision medicine tests, breast cancer has been an early success story, with several genomic assays already available as treatment decision-making aids. In contrast, no published intervention has used health services big data resources to improve breast cancer care, but templates are available for such efforts in other cancer types. For example, the Michigan Urological Surgery Improvement Collaborative has effectively leveraged EHR and claims data together with other data resources to directly inform providers about practice patterns, improve adherence to guidelines, and optimize patient treatment.[42,43] In North Carolina, the big data infrastructure of the ICISS has been used to identify or verify “hot spots” of low rates of colorectal cancer screening and high racial disparities in morbidity and mortality. These hotspotting data were used to simulate the effect of different intervention scenarios, which led to an ongoing intervention specifically aimed to reduce disparities in screening.[44,45] The ongoing ACCURE (Accountability for Cancer Care Through Undoing Racism and Equity) intervention uses an electronic alert system linked to the EHR, along with patient navigation and community-based participatory research, to decrease disparities between black patients and white patients in terms of receipt of timely care for breast and lung cancer; results have not yet been reported.[46] A critical component of successful interventions to address disparities in cancer care is a strong interdisciplinary approach to the entire “lifecycle” of the project (Figure 2). Clinical oncology experts must work in close harmony with data and software engineers to store and manage the data. Additionally, the research scientists and analysts coaxing valuable and actionable information from these large systems need to have close working relationships and intimate knowledge of the clinical data processes that generated their analytic files. Integration of biological data and access data across multiple scales also requires a team approach that employs a wide range of expertise. Exploiting the power of the rich breast cancer disparities data available will require teams that have collective expertise in the areas of clinical oncology, computer science, molecular biology, statistical analysis, and population science, and that can flexibly tackle new methodological challenges as they arise.
Financial Disclosure: Dr. Meyer is a data and methods consultant to, and serves on the advisory board of, Merck. The other authors have no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
References:
1. DeSantis CE, Fedewa SA, Goding Sauer A, et al. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31-42.
2. Ward JS, Barker A. Undefined by data: a survey of big data definitions 2013. May 30, 2017. https://arxiv.org/abs/1309.5821. Accessed July 13, 2017.
3. Kayyali B, Knott D, Van Kuiken S. The big-data revolution in US health care: accelerating value and innovation: McKinsey & Company; 2013. http://www.mckinsey.com/industries/healthcare-systems-and-services/our-insights/the-big-data-revolution-in-us-health-care. Accessed July 13, 2017.
4. De Mauro A, Greco M, Grimaldi M. A formal definition of big data based on its essential features. Library Rev. 2016;65:122-35.
5. Daly B, Olopade OI. A perfect storm: how tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J Clin. 2015;65:221-38.
6. Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492-502.
7. Howlader N, Altekruse SF, Li CI, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. 2014;106.
8. O’Brien KM, Cole SR, Tse CK, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100-10.
9. Troester MA, Sun X, Allott EH, et al. Racial differences in PAM50 subtypes in the Carolina Breast Cancer Study. J Natl Cancer Inst. 2017 Aug 1. [Epub ahead of print]
10. Huo D, Hu H, Rhie SK, et al. Comparison of breast cancer molecular features and survival by African and European ancestry in The Cancer Genome Atlas. JAMA Oncol. 2017 May 4. [Epub ahead of print]
11. Freedman RA, He Y, Winer EP, et al. Trends in racial and age disparities in definitive local therapy of early-stage breast cancer. J Clin Oncol. 2009;27:713-9.
12. Reeder-Hayes KE, Bainbridge J, Meyer AM, et al. Race and age disparities in receipt of sentinel lymph node biopsy for early-stage breast cancer. Breast Cancer Res Treat. 2011;128:863-71.
13. Carpenter WR, Reeder-Hayes K, Bainbridge J, et al. The role of organizational affiliations and research networks in the diffusion of breast cancer treatment innovation. Med Care. 2011;49:172-9.
14. Meyer AM, Reeder-Hayes KE, Liu H, et al. Differential receipt of sentinel lymph node biopsy within practice-based research networks. Med Care. 2013;51:812-8.
15. Freedman RA, Kouri EM, West DW, et al. Racial/ethnic differences in patients’ selection of surgeons and hospitals for breast cancer surgery. JAMA Oncol. 2015;1:222-30.
16. Wheeler SB, Carpenter WR, Peppercorn J, et al. Structural/organizational characteristics of health services partly explain racial variation in timeliness of radiation therapy among elderly breast cancer patients. Breast Cancer Res Treat. 2012;133:333-45.
17. Hershman DL, Wang X, McBride R, et al. Delay in initiating adjuvant radiotherapy following breast conservation surgery and its impact on survival. Int J Radiat Oncol Biol Phys. 2006;65:1353-60.
18. Fedewa SA, Ward EM, Stewart AK, et al. Delays in adjuvant chemotherapy treatment among patients with breast cancer are more likely in African American and Hispanic populations: a national cohort study 2004-2006. J Clin Oncol. 2010;28:4135-41.
19. Hershman D, McBride R, Jacobson JS, et al. Racial disparities in treatment and survival among women with early-stage breast cancer. J Clin Oncol. 2005;23:6639-46.
20. Freedman RA, Hughes ME, Ottesen RA, et al. Use of adjuvant trastuzumab in women with human epidermal growth factor receptor 2 (HER2)-positive breast cancer by race/ethnicity and education within the National Comprehensive Cancer Network. Cancer. 2013;119:839-46.
21. Reeder-Hayes K, Peacock Hinton S, Meng K, et al. Disparities in use of human epidermal growth hormone receptor 2-targeted therapy for early-stage breast cancer. J Clin Oncol. 2016;34:2003-9.
22. Roberts MC, Wheeler SB, Reeder-Hayes K. Racial/ethnic and socioeconomic disparities in endocrine therapy adherence in breast cancer: a systematic review. Am J Public Health. 2015;105(suppl 3):e4-e15.
23. Roberts MC, Weinberger M, Dusetzina SB, et al. Racial variation in the uptake of Oncotype DX testing for early-stage breast cancer. J Clin Oncol. 2016;34:130-8.
24. Davis BA, Aminawung JA, Abu-Khalaf MM, et al. Racial and ethnic disparities in Oncotype DX test receipt in a statewide population-based study. J Natl Compr Canc Netw. 2017;15:346-54.
25. Meyer AM, Olshan AF, Green L, et al. Big data for population-based cancer research: the integrated cancer information and surveillance system. N C Med J. 2014;75:265-9.
26. Truven Health Analytics; IBM Watson Health. Putting research data into your hands with the MarketScan databases. 2017 [cited May 25, 2017]. http://truvenhealth.com/markets/life-sciences/products/data-tools/marketscan-databases. Accessed July 11, 2017.
27. Kent EE, Malinoff R, Rozjabek HM, et al. Revisiting the Surveillance Epidemiology and End Results Cancer Registry and Medicare Health Outcomes Survey (SEER-MHOS) linked data resource for patient-reported outcomes research in older adults with cancer. J Am Geriatr Soc. 2016;64:186-92.
28. Chubak J, Yu O, Pocobelli G, et al. Administrative data algorithms to identify second breast cancer events following early-stage invasive breast cancer. J Natl Cancer Inst. 2012;104:931-40.
29. Chawla N, Yabroff KR, Mariotto A, et al. Limited validity of diagnosis codes in Medicare claims for identifying cancer metastases and inferring stage. Ann Epidemiol. 2014;24:666-72.
30. Bastien RR, Rodriguez-Lescure A, Ebbert MT, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics. 2012;5:44.
31. Allott EH, Cohen SM, Geradts J, et al. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER consortium. Cancer Epidemiol Biomarkers Prev. 2016;25:470-8.
32. Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra13.
33. Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929-44.
34. Keenan T, Moy B, Mroz EA, et al. Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol. 2015;33:3621-7.
35. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415-21.
36. Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979-93.
37. Shah A, Stewart AK, Kolacevski A, et al. Building a rapid learning health care system for oncology: why CancerLinQ collects identifiable health information to achieve its vision. J Clin Oncol. 2016;34:756-63.
38. Berger ML, Curtis MD, Smith G, et al. Opportunities and challenges in leveraging electronic health record data in oncology. Future Oncol. 2016;12:1261-74.
39. Penn DC, Chang Y, Meyer AM, et al. Provider-based research networks may improve early access to innovative colon cancer treatment for African Americans treated in the community. Cancer. 2015;121:93-101.
40. Carpenter WR, Meyer AM, Wu Y, et al. Translating research into practice: the role of provider-based research networks in the diffusion of an evidence-based colon cancer treatment innovation. Med Care. 2012;50:737-48.
41. Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care.
J Clin Oncol. 2010;28:4268-74.
42. Ross I, Womble P, Ye J, et al. MUSIC: patterns of care in the radiographic staging of men with newly diagnosed low risk prostate cancer. J Urol. 2015;193:1159-62.
43. Hurley P, Dhir A, Gao Y, et al. A statewide intervention improves appropriate imaging in localized prostate cancer. J Urol. 2017;197:1222-8.
44. Wheeler SB, Kuo TM, Goyal RK, et al. Regional variation in colorectal cancer testing and geographic availability of care in a publicly insured population. Health Place. 2014;29:114-23.
45. Wheeler SB, Kuo TM, Meyer AM, et al. Multilevel predictors of colorectal cancer testing modality among publicly and privately insured people turning 50. Prev Med Reports. 2017;6:9-16.
46. Schaal JC, Lightfoot AF, Black KZ, et al. Community-guided focus group analysis to examine cancer disparities. Progress in community health partnerships: research, education, and action. Prog Community Health Partnersh. 2016;10:159-67.