Artificial Intelligence in Oncology: Current Applications and Future Directions
In this review, we introduce the fundamentals of artificial intelligence and provide an overview of its current applications, pitfalls, and future directions in oncology.
Oncology (Williston Park). 33(2):46-53.

Benjamin H. Kann, MD

Reid Thompson, MD, PhD

Charles R. Thomas, Jr, MD

Adam Dicker, MD, PhD

Sanjay Aneja, MD

Figure 1. Relationship Between Artificial Intelligence, Machine Learning, and Deep Learning
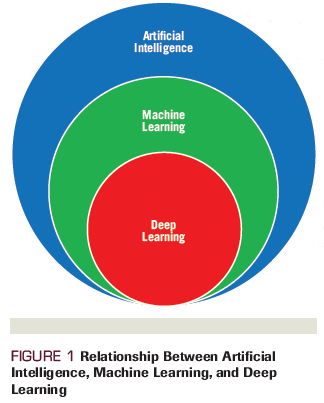
Figure 2. Neurologic Basis of Neural Network Architectures
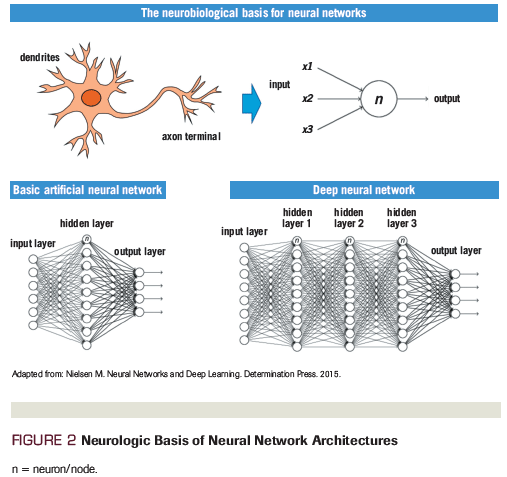
Figure 3. Rise in Deep Learning Publications
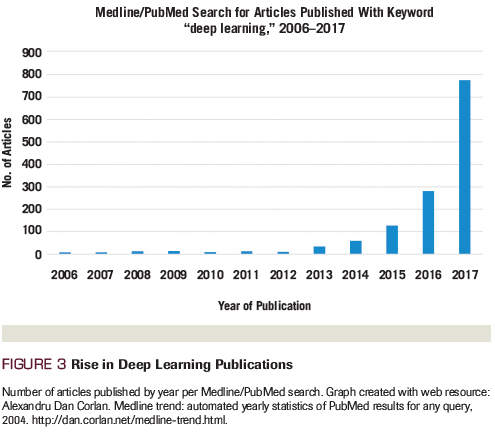
Table 1. Applications of Artificial Intelligence in Imaging
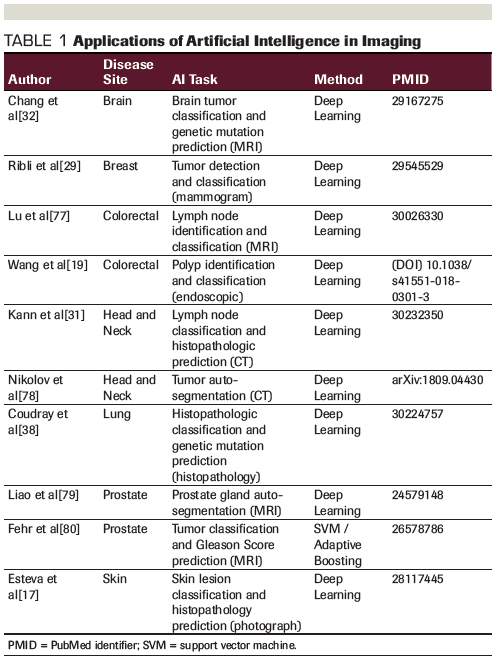
Table 2. Applications of Artificial Intelligence in Clinical Outcome Prediction
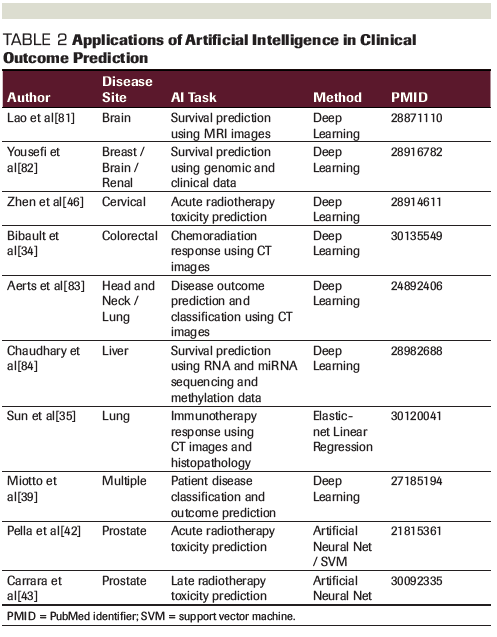
Table 3. Applications of Artificial Intelligence in Translational Oncology
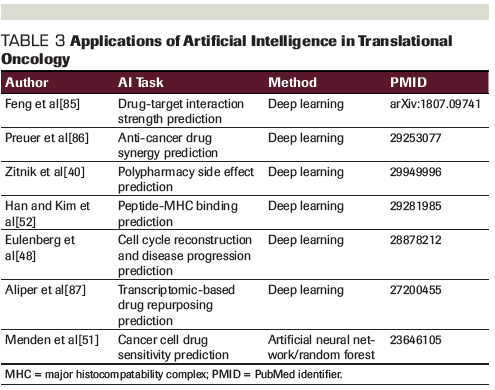
Artificial Intelligence: Terms and Definitions
Click here to read an expert perspective from Tufia C. Haddad, MD.
Introduction
Artificial intelligence (AI), for years, has captured society’s imagination and generated enthusiasm for its potential to improve our lives. Presently, AI already plays an integral role in our daily routines and our interactions with media, transportation, and communications. There is an increasing interest in the applications of AI in healthcare to improve disease diagnosis, management, and the development of effective therapies. Given the large number of patients diagnosed with cancer and significant amount of data generated during cancer treatment, there is a specific interest in the application of AI to improve oncologic care. In this review, we introduce the fundamentals of AI and provide an overview of its current applications, pitfalls, and future directions in oncology.
What is AI?
Originally formalized in the 1950s, AI refers to the ability of a machine to perform tasks commonly associated with intelligent human behavior.[1] Including disciplines from both computer science and mathematics, AI can be considered a group of iterative, "self-learning" techniques, which discover relationships within data that can evolve and often be performed faster over time.
Machine Learning (ML): The Engine That Drives AI
A majority of the AI applications within healthcare involve the utilization of ML algorithms. As ML algorithms are exposed to more training data, they are able to appreciate hidden patterns within the data which can then be used to perform a task without explicit programming.[2] There are dozens of ML algorithms that have been proposed over the past several decades, and the most traditional forms of ML, such as logistic regression, have proven themselves as valuable tools for general clinical oncology research.[3]
ML tasks are often broadly dichotomized into supervised or unsupervised learning. In supervised learning, a labeled dataset of inputs and outputs is used to train the ML algorithm. The algorithm attempts to learn a general rule that maps input to output. Supervised ML algorithms can learn patterns in data for categorical outputs (classification) and continuous data (regression). Conversely, unsupervised ML algorithms use unlabeled data, with the goal of discovering structure in the input data. Unsupervised ML algorithms are often used to simplify (dimensionality reduction) or organize (clustering) data.
In traditional ML tasks, there is often some pre-engineered organization of raw input data into features that are inferred to have an impact on output.
Artificial Neural Networks and Deep Learning (DL): The Wave Comes to Healthcare
One drawback of traditional ML algorithms has been the need for pre-engineered organization of raw input data into structured datasets. The inability of certain ML algorithms to use unstructured data from the point of generation has limited their utility in clinical practice. One ML algorithm well suited to analyze unstructured data is known as DL (Figure 1).
DL algorithms are often synonymous with AI. DL is a form of ML that uses layered "artificial neural networks" to develop sophisticated models with the ability to understand data at different levels of abstraction.[4] Artificial neural networks (ANNs) were inspired by human neurobiology and the ability of the brain to use cascading, varying, and layered combinations of neurons to learn complicated patterns with a hierarchy of progressively more complex features (Figure 2). Modeling the human neuron in computers yielded the basic design of early ANNs.[5]
While traditional ML algorithms used pre-engineered features to develop predictions, DL algorithms are able to learn the optimal features that best fit the data through the training process, avoiding the need to use pre-engineering, unstructured data. This ability has allowed DL algorithms to outperform traditional ML algorithms in many common AI problems, including image classification, natural language processing, and sequence prediction.[4,6,7] Accordingly, the number of research articles published involving deep learning in the medical field has skyrocketed over the past few years (Figure 3).
Due to this DL renaissance, AI has now generated significant attention in healthcare.[8-11] In oncology, DL has become a natural partner in the pursuit of precision medicine,[12-15] leveraging vast, heterogenous datasets to better diagnosis disease burden, predict patient outcomes, and tailor management. AI can also be coupled to a multitude of emerging mobile health interfaces, such as smartphone apps and wearable devices, to develop “digital biomarkers” that can explain, influence, and predict clinical outcomes. In the following sections, we will delve into current AI applications in oncology, their limitations, and future implications.
AI and Cancer Imaging
Seeing better with convolutional neural networks
Image analysis has proven to be among the most effective methods in which AI has impacted society. AI powered by DL algorithms has provided us self-driving vehicles, mobile check deposit, and multiple other useful technologies. Given the large amount of digital imaging data present within medicine, there is increasing excitement about the application of similar techniques to imaging within oncology (Table 1).
This revolution in image analysis was catalyzed by the development of a particular DL architecture, the Convolutional Neural Network (CNN). CNNs analyze pixel-level information from images. The added benefit of CNNs compared to other DL configurations is the ability to account for orientation of the pixels in relation to one another. This effectively allows the CNN to appreciate lines, curves, and eventually objects within images. CNN-based models have recently been shown to be equivalent to humans in picture classification and object detection.[6,16]
Clinical photographs
One of the initial papers highlighting the promise of DL in cancer imaging was in identification of skin cancer based on skin photographs.[17] The CNN trained on 130,000 skin images was able to classify malignant lesions with higher sensitivity and specificity than a panel of 21 board-certified dermatologists. Practical applications of detecting skin pathology utilizing patients as the generator of imaging input data has evolved.[18] Further use of CNNs to classify digital photography has been in the automatic detection of polyps during colonoscopy. One particular study highlights that CNNs can be used for not only image classification, but also to detect regions of clinical importance. Researchers found that CNN, which was trained on colonoscopic images from 1,290 patients, had a 94% sensitivity in polyp detection.[19]
Radiographic imaging
Given one of the most effective applications of AI techniques has been in the field of computer vision, there is naturally excitement in the field of radiology, where there exist a number of digitized images. The goals of these AI algorithms have ranged from assisted diagnosis to outcome prediction.
AI algorithms have been found to be effective in streamlining cancer screening and detection. Automated lung nodule detection and classification has attracted significant attention and formed the basis of the 2017 Kaggle Data Science Bowl, an international competition for ML scientists.[20] Several successful CNN-based models resulted from this competition and other research groups, demonstrating accuracies in the 80% to 95% range and showing promise for lung cancer screening.[21–26] Additionally, CNNs have been shown to be successful in segmenting tumor volumes, which may have implications for radiotherapy treatment planning.[27] Improvement of breast cancer screening with AI has also been an active area of investigation, with its own data science competitions,[28] resulting in a CNN algorithm able to detect breast malignancy with a sensitivity of 90%.[29,30]
AI has also shown promise in detecting radiographic anatomic features of malignancies that goes beyond what can be reliably achieved by clinicians. While extranodal extension (ENE) of tumors in the head and neck cancer lymph nodes has been notoriously difficult to diagnosis radiographically by clinicians, a CNN-based model showed >85% accuracy in identifying this feature on diagnostic, contrast-enhanced CT scans.[31] Identification of ENE has high importance in prognostication and management for head and neck cancer patients, and thus this model shows promise as a clinical decision-making tool.
Going further than anatomic characterization, AI has shown promise in the burgeoning field of radiogenomics, where radiographic image analysis is used to predict underlying genotypic traits. This has recently been demonstrated using CNNs on brain MRIs of patients with low-grade glioma. CNNs have been able to predict both IDH mutation and MGMT methylation status with 85% to 95% and 83% accuracy, respectively, based on raw imaging data alone.[32,33]
DL also has the potential to predict response to treatment based on imaging findings. Recently, a CNN model showed success in predicting complete response to neoadjuvant chemoradiation with 80% accuracy.[34] Additionally, a radiomics signature using extracted features from CT data and a ML algorithm were able to predict underlying CD8 cell tumor infiltration and, remarkably, response to immunotherapy for a variety of advanced cancers.[35]
Digital pathology
The increasing digitization of histopathologic tumor specimen slides provides a robust 2D image suitable for DL analysis. DL CNN algorithms have now been shown to diagnose breast cancer metastasis in lymph nodes with at least equivalent performance compared to a panel of pathologists, and in a more time-efficient manner.[36] DL has also been shown to be useful in automated Gleason grading of prostate adenocarcinoma Hematoxylin and Eosin–stained specimens, with a 75% rate of agreement between the algorithm and pathologists.[37]
DL algorithms have also gone a step further than pathologic diagnosis automation and have been used to characterize the underlying genotype-phenotype correlation within a tumor specimen. Using raw input data consisting of digitized formalin-fixed, paraffin-embedded tissue from lung cancer biopsies, a CNN was trained to predict six different genetic mutations (STK11, EGFR, FAT1, SETBP1, KRAS, and TP53), establishing that genotypic information may be gleaned from histopathologic architectural patterns.[38] These methods may assist pathologists in the detection of cancer gene mutations and may be cost-effective compared to direct mutational analysis.
AI and Clinical Outcomes
Within clinical oncology, AI has increasingly been applied to harness the power of the electronic health record (EHR) (Table 2). Specifically, AI-based natural language processing techniques have shown promise in predicting the development of diseases across large healthcare systems. A notable example from a group at Mount Sinai, a DL-based AI algorithm modeling EHR, was able to predict the development of a variety of diseases with 93% accuracy overall, including cancers of the prostate, rectum, and liver.[39]
There has additionally been interest in using DL to predict cancer treatment toxicity. Recently, a CNN approach was used to predict side effects of polypharmacy combinations based on databases of protein-protein and drug-protein interactions.[40] This study led to the discovery of at least five novel drug-drug interaction predictions, which were subsequently found to have supporting literature evidence. Use of AI to predict radiotherapy toxicity has generated significant interest over the past few years.[41] Basic neural networks, CNNs, and other ML methods have been explored, using clinical and dosimetric data to predict urinary and rectal toxicity resulting from prostate radiotherapy results,[42–44] hepatobiliary toxicity after liver radiotherapy,[45] and rectal toxicity for patients receiving radiotherapy for cervical cancer.[46]
AI and Translational Oncology
Translational oncology is an area where AI is beginning to emerge. Over the past decade, there has been an expansion of biological quantitative or “-omic” data. Given the complexities and heterogeneity within this data, the use of DL in analysis has been appealing. DL neural networks have been utilized to predict protein structure,[47] classify cells into a distinct stage of mitosis,[48] and even predict the future lineage of progenitor cells based on microscopy images.[49]
Drug development and repurposing has become an attractive target for DL. One group used DL ANNs trained on transcriptomic response signatures to drugs to predict with high accuracy the likelihood of failure of a clinical trial of over 200 example drugs.[50] Another used an ANN to predict cancer cell sensitivity to therapeutics using a combination of genomic and chemical properties.[51] CNNs have also been employed to predict peptide-major histocompatibility complex binding,[52] which may have implications for oncologic immunotherapy development. See Table 3 for a summary of applications of AI in translational oncology that have been researched.
AI and Clinical Decision Making
Given the increasing amount and pace of published research, clinical trial enrollment, drug development, and biomarker discovery in oncology in recent years, there is more opportunity than ever for AI to assist in synthesizing this data and to guide decision-making. There are several commercial applications in development that utilize DL and natural language processing to this aim.[53] These applications are being designed to link patient data to clinical trial databases and to match patients to appropriate clinical trials nationwide. Another algorithm utilizes ML to select the appropriate investigational drugs in development for a given patient. There has also been interest in utilizing AI coupled with patient data and national treatment guidelines to guide cancer management, with the most prominent example being IBM’s Watson for Oncology (WFO). WFO has demonstrated high concordance with tumor board recommendations for breast cancer patients, though has fallen short in other areas of oncologic decision-making.[54,55] While this area of AI application is in its nascent stages, performance continues to improve, and there is great potential to improve clinical practice.
AI Limitations and Future Directions
Proving generalizability and real-world applications
While AI is rapidly being incorporated into oncologic research, work remains to be done to translate these studies into real-world, clinically meaningful applications. One of the biggest barriers is in external validation and proving generalizability of DL applications. Given the complexity of neural networks and the extremely large number of parameters (often in the millions), there is a high tendency for neural networks to create overfitted models that do not generalize across different populations. Additionally, because there is a significant amount of heterogeneity of medical data across institutions, multiple external validation sets may be required to prove the performance of an application.[56]
Data access and equity
Directly contributing to this problem of overfitting are limitations with data access and quality. DL neural networks, more than any other ML algorithm, require large amounts of data. This can pose a problem in healthcare when attempting to apply AI to disease processes with less prevalence. Furthermore, data is often siloed within individual institutions. Contributing to this relative data drought are concerns with transmission of protected patient health information, along with lack of data-sharing infrastructure to link institutions, heterogeneity and incompleteness in the collection of data, and competition between institutions. These obstacles are beginning to be addressed, with more and more emphasis on streamlined data capture,[57] and a number of multi-institutional data-sharing agreements[58-61] Guidelines have been proposed to support FAIR (findable, accessible, interoperable, reusable) data use,[62] and there are now opportunities for research groups to publish their data itself, which may incentivize openness.[63]
Interpretability and the black box problem
One of the central limitations to adoption of AI in healthcare is the concern that these models, despite regularly achieving high performance, are somewhat opaque. For instance, a DL model may correctly predict that a patient will develop pancreatic cancer based on his past 2 years of EHR data, but why did it make that prediction? At the present, we are limited in our ability to determine the precise logic behind DL-based predictions. This is often referred to as the “black box” problem.[64] In medical practice, it has traditionally been essential in clinical decision-making to know the rationale for each decision. Traditional ML algorithms, like linear regression, have limited ability to model complex relationships, but offer this easy interpretability-in these algorithms, we have a set of pre-defined features and the resulting feature weights that characterize their effect sizes. In contrast, DL utilizes unstructured input data, and the bulk of knowledge generation occurs within the hidden layers. It thus becomes difficult to determine which specific characteristic(s) of the input data contributed to the outcome. This interpretability challenge has large implications for adoption of AI-based algorithms in healthcare, both from practitioner and regulatory perspectives.[65-68]
Tackling the black box problem has now become a major focus of research.[69] In AI image analysis algorithms, several methods have been developed, including feature visualization, saliency maps, class activation mapping, and sensitivity analyses, where certain parts of the image are hidden to the effect on prediction.[70] While these methods have advanced over the past few years, further work is needed to better elucidate the decision-making logic with deep neural networks.
Education and expertise
To successfully merge AI with clinical oncology and maximize its impact, there are knowledge gaps that need to be addressed. Currently, physicians receive little training in data science and ML, limiting their ability to understand DL mechanisms, adopt algorithms appropriately, and conduct research.[71] Similarly, most data scientists have little experience with oncologic workup and management, limiting the ability to identify important and suitable clinical use cases. Further collaboration should be pursued between clinical oncologic departments and bioinformatics and data science divisions, and strategic partnerships with technology firms should be formed where appropriate.
Promoting AI in Oncology: Professional Societies and National Initiatives
In response to these challenges, several national professional societies have launched initiatives to bridge these knowledge gaps and promote the dissemination of AI in oncology.
American College of Radiology (ACR)
The ACR has founded a Data Science Institute (ACR-DSI) with the mission of collaborating with radiologists, industry, and government agencies to facilitate the development of AI in imaging.[72] Within the ACR-DSI are several core goals: 1) providing standards for measuring performance of AI algorithms (“Touch-AI”), 2) independent, external validation of algorithms and navigating the regulatory landscape (“Certify-AI”), and longitudinal, prospective evaluation of deployed algorithm performance “(Assess-AI”). The ACR-DSI has additionally set up a series of use cases for recommended AI imaging applications with unmet clinical need.
American Society of Clinical Oncology (ASCO) and American Society for Radiation Oncology (ASTRO)
ASCO has launched a big data initiative named CancerLinQ, in partnership with oncologists, industry, and academia, with the goals of real-time quality of care tracking and treatment evaluation, as well as knowledge dissemination to oncologists in user-friendly ways.[73] The initiative’s backbone is a constantly growing database of de-identified patient information that can be mined and analyzed. In 2017, ASTRO partnered with CancerLinQ to provide radiation oncology expertise and uses for the database. In addition, Big Data Analytics and Bioinformatics is one of the core initiatives of the ASTRO Research Agenda for 2018.[74]
National Institutes of Health (NIH)
As part of the NIH Common Fund, the Big Data to Knowledge (BD2K) initiative was launched to support the research and development of tools to integrate big data and data science into biomedical research.[75] One of the central components of the initiative involves leveraging existing national datasets, such as the Library of Integrated Network-based Cellular Signatures (LINCS) and The Cancer Genome Atlas (TCGA), and applying ML techniques to discover patterns in the data that may result in heretofore unknown compounds for cancer therapeutics.[76]
Conclusions
Over the past decade, AI has undergone a reawakening. Due to an explosion of electronic data, advances in technological infrastructure, and groundbreaking research in DL neural networks, AI is poised to make practice-changing impacts on the medical field and oncologic care. At present, AI has shown promise in improving cancer imaging diagnostics and treatment response evaluation, predicting clinical outcomes, and catalyzing drug development and translational oncology. Obstacles remain-such as validation and proving generalizability, concerns over interpretation, and the widening knowledge gap between clinical and data science experts. If we can address these challenges, AI has the potential to transform oncology, harnessing the power of big data to drive cancer care into the 21st century and beyond.
Financial Disclosure: The authors have no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
PERSPECTIVE
Artificial Intelligence as a Solution to Burnout in Oncology
Tufia C. Haddad, MD
Developing applications of artificial intelligence (AI) and cognitive systems in oncology requires a collaborative, multidisciplinary effort that extends far beyond medicine and computer science. More than a few significant challenges, however, limit the translation of AI-related cancer research into meaningful clinical applications. Among these are the abilities to form partnerships across multiple industries, to gain equitable access to large volumes of annotated data, and to conduct unbiased training of machine-learning algorithms. The article by Aneja and colleagues provides a detailed overview of the evolving field of AI in oncology and serves as a primer for curious and reluctant oncologists alike, the vast majority of whom have not received formal training in data science.
While there has been hype that AI has the potential to replace doctors, early experience has suggested that AI can instead augment human intelligence, enabling doctors to perform with greater efficiency, engagement, and effectiveness. That said, a more credible, contemporary threat faces the oncology profession: unprecedented levels of burnout.[1] Although multiple factors have been associated with this state of stress, the ubiquitous involvement of computers in all aspects of medicine is recognized as a major driver. For example, while electronic health records (EHR), computerized prescribing, and order entry were established to improve quality and coordination of patient care, implementation of these systems has resulted in unintended consequences for providers: reduced efficiency and increased clerical and cognitive burden.[2] During office hours, physicians spend almost 2 hours engaged in EHR and desk work for every 1 hour of direct patient face time,[3] and it is estimated that an additional 1.4 hours of EHR interactions occur daily outside of business hours.[4]
The EHR contains an enormous volume of data, and new data sources have the potential to generate a tsunami of additional patient data points. These include tumor genome sequencing reports, electronic patient-reported outcomes responses, patient online messaging, and patient-generated health data from apps and wearables. Integration of these data within the EHR and clinical workflows is currently suboptimal or lacking for most practices, furthering the rift between providers and their computers. Concurrent with this explosion of data is the frenzied pace of knowledge gains in oncology, including a record of 51 new agents or new indications for existing agents that were approved by the US Food and Drug Administration for cancer treatment in 2018.[5]
It has been recognized that much of the existing digital workload can be delegated to other care team members or to data abstraction and documentation specialists. Alternatively, technology may be exactly what is needed to solve our technology problem. Specifically, natural language processing (NLP), a branch of AI, can interpret, augment, and transform free text so that it can be represented for computation.[6] In medicine, it leverages the EHR, including its most valuable asset: the clinical note. NLP serves as the engine for cognitive systems, but adoption of this technology requires expert training and supervision, as well as thoughtful implementation, with minimal disruption to clinical workflows.
Under these conditions, AI offers the potential for detailed EHR summarization in a single click, as well as clinical decision support at the point of care aligned with evidence-based care pathways. AI may also generate order sets, cancer registries, or personalized care plans for treatment or survivorship. Finally, it may provide optimized billing codes and quality/outcomes reporting for programmatic assessments and regulatory mandates. While extraordinary investments have aimed to create AI applications for cancer risk prediction, diagnosis, and treatment, perhaps an equally noble goal would be to develop the NLP capabilities of cognitive systems to eradicate the growing mindless hours spent navigating the EHR, thereby allowing oncologists to do what they do best: provide the highest level of cutting-edge, patient-centered care to those facing cancer.
FINANCIAL DISCLOSURE: Dr. Haddad has no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
REFERENCES
1. Shanafelt TD, Gradishar WJ, Kosty M, et al. Burnout and career satisfaction among US oncologists. J Clin Oncol. 2014;32:678-86.
2. Shanafelt TD, Dyrbye LN, Sinsky C, et al. Relationship between clerical burden and characteristics of the electronic environment with physician burnout and professional satisfaction. Mayo Clin Proc. 2016;91:836-48.
3. Sinsky C, Colligan L, Li L, et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialities. Ann Intern Med. 2016;165:753-60.
4. Arndt BG, Beasley JW, Watkinson MD, et al. Tethered to the EHR: primary care physician workload assessment using EHR event log data and time-motion observations. Ann Fam Med. 2017;15:419-26.
5. U.S. Food & Drug Administration. Hematology/oncology (cancer) approvals & safety notifications. https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm279174.htm. Updated January 15, 2019. Accessed January 25, 2019.
6. Yim WW, Yetisgen M, Harris WP, et al. JAMA Oncol. 2016;2:797-804.
Dr. Haddad is a Medical Oncologist, Chair of the Breast Medical Oncology practice, and Chair of Health Information Technology in the Department of Oncology at the Mayo Clinic in Rochester, Minnesota.
References:
1. Russell SJ, Norvig P. Artificial intelligence: a modern approach. 3rd edition. Upper Saddle River, New Jersey: Prentice Hall; 2003.
2. Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3:210-29.
3. Kourou K, Exarchos TP, Exarchos KP, et al. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8-17.
4. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-44.
5. Hopfield JJ. Artificial neural networks. IEEE Circuits Devices Mag. 1988;4:3-10.
6. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing systems 25. Curran Associates, Inc; 2012. pp. 1097–105. http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. Accessed March 15, 2018.
7. Hirschberg J, Manning CD. Advances in natural language processing. Science. 2015;349:261-6.
8. Hinton G. Deep learning: a technology with the potential to transform health care. JAMA. 2018;320:1101-2.
9. Mukherjee S. A.I. versus M.D. The New Yorker. March 2017. https://www.newyorker.com/magazine/2017/04/03/ai-versus-md. Accessed December 3, 2018.
10. Singer N. How big tech is going after your health care. The New York Times. December 27, 2017. https://www.nytimes.com/2017/12/26/technology/big-tech-health-care.html. Accessed December 3, 2018.
11. Mims C. The AI doctor will see you now. The Wall Street Journal. May 20, 2018. https://www.wsj.com/articles/the-ai-doctor-will-see-you-now-1526817600. Accessed December 3, 2018.
12. Fröhlich H, Balling R, Beerenwinkel N, et al. From hype to reality: data science enabling personalized medicine. BMC Med. 2018;16:150.
13. Bibault JE, Giraud P, Burgun A. Big data and machine learning in radiation oncology: state of the art and future prospects. Cancer Lett. 2016;382:110-7.
14. Shrager J, Tenenbaum JM. Rapid learning for precision oncology. Nat Rev Clin Oncol. 2014;11:109-18.
15. Ding MQ, Chen L, Cooper GF, et al. Precision oncology beyond targeted therapy: combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Mol Cancer Res. 2018;16:269-78.
16. Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211-52.
17. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-8.
18. Webster DE, Suver C, Doerr M, et al. The Mole Mapper Study, mobile phone skin imaging and melanoma risk data collected using ResearchKit. Sci Data. 2017;4:170005.
19. Wang P, Xiao X, Brown JRG, et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741.
20. Data Science Bowl 2017. https://www.kaggle.com/c/data-science-bowl-2017. Accessed December 3, 2018.
21. de Wit J. 2nd place solution for the 2017 National Data Science Bowl. http://juliandewit.github.io/kaggle-ndsb2017/. Accessed March 15, 2018.
22. Hammack D. DSB2017: Code for 2nd place solution to the 2017 National Data Science Bowl; 2018. https://github.com/dhammack/DSB2017. Accessed January 12, 2018.
23. Kuan K, Ravaut M, Manek G, et al. Deep learning for lung cancer detection: tackling the Kaggle Data Science Bowl 2017 challenge. May 2017. http://arxiv.org/abs/1705.09435. Accessed February 14, 2019.
24. Zhao W, Yang J, Sun Y, et al. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018;78:6881-9.
25. Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep. 2017;7:46479.
26. Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLOS One. 2017;12:e0188290.
27. Wang S, Zhou M, Gevaert O, et al. A multi-view deep convolutional neural networks for lung nodule segmentation. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:1752-5.
28. Sage Bionetworks. Digital Mammography DREAM Challenge. http://sagebionetworks.org/research-projects/digital-mammography-dream-challenge/. Accessed December 3, 2018.
29. Ribli D, Horváth A, Unger Z, et al. Detecting and classifying lesions in mammograms with deep learning. Sci Rep. 2018;8:4165.
30. Trister AD, Buist DSM, Lee CI. Will machine learning tip the balance in breast cancer screening? JAMA Oncol. 2017;3:1463-4.
31. Kann BH, Aneja S, Loganadane GV, et al. Pretreatment identification of head and neck cancer nodal metastasis and extranodal extension using deep learning neural networks. Sci Rep. 2018;8:14036.
32. Chang K, Bai HX, Zhou H, et al. Residual convolutional neural network for determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073-81.
33. Chang P, Grinband J, Weinberg BD, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39:1201-7.
34. Bibault JE, Giraud P, Durdux C, et al. Deep learning and radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611.
35. Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180-91.
36. Bejnordi BE, Veta M, van Diest PJ, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199-210.
37. Arvaniti E, Fricker KS, Moret M, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8:12054.
38. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559-67.
39. Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016;6:26094.
40. Zitnik M, Agrawal M, Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34:i457-i466.
41. Kang J, Schwartz R, Flickinger J, Beriwal S. Machine learning approaches for predicting radiation therapy outcomes: a clinician’s perspective. Int J Radiat Oncol. 2015;93:1127-35.
42. Pella A, Cambria R, Riboldi M, et al. Use of machine learning methods for prediction of acute toxicity in organs at risk following prostate radiotherapy. Med Phys. 2011;38:2859-67.
43. Carrara M, Massari E, Cicchetti A, et al. Development of a ready-to-use graphical tool based on artificial neural network classification: application for the prediction of late fecal incontinence after prostate cancer radiation therapy. Int J Radiat Oncol. 2018 Aug 6. [Epub ahead of print]
44. Lee S, Kerns S, Ostrer H, et al. Machine learning on a genome-wide association study to predict late genitourinary toxicity after prostate radiation therapy. Int J Radiat Oncol Biol Phys. 2018;101:128-35.
45. Ibragimov B, Toesca D, Chang D, et al. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45:4763-74.
46. Zhen X, Chen J, Zhong Z, et al. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Phys Med Biol. 2017;62:8246-63.
47. Wang J, Cao H, Zhang JZH, Qi Y. Computational protein design with deep learning neural networks. Sci Rep. 2018;8:6349.
48. Eulenberg P, Köhler N, Blasi T, et al. Reconstructing cell cycle and disease progression using deep learning. Nat Commun. 2017;8:463.
49. Buggenthin F, Buettner F, Hoppe PS, et al. Prospective identification of hematopoietic lineage choice by deep learning. Nat Methods. 2017;14:403-6.
50. Artemov AV, Putin E, Vanhaelen Q, et al. Integrated deep learned transcriptomic and structure-based predictor of clinical trials outcomes. December 29, 2016. https://www.biorxiv.org/content/10.1101/095653v2.
51. Menden MP, Iorio F, Garnett M, et al. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLOS One. 2013;8:e61318.
52. Han Y, Kim D. Deep convolutional neural networks for pan-specific peptide-MHC class I binding prediction. BMC Bioinformatics. 2017;18:585.
53. Sennaar K. AI and machine learning for clinical trials: examining 3 current applications. Emerj - Artificial Intelligence Research and Insight. https://emerj.com/ai-sector-overviews/ai-machine-learning-clinical-trials-examining-x-current-applications/. Accessed January 18, 2019.
54. Somashekhar SP, Sepúlveda MJ, Puglielli S, et al. Watson for oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board. Ann Oncol. 2018;29:418-23.
55. Liu C, Liu X, Wu F, et al. Using artificial intelligence (Watson for Oncology) for treatment recommendations amongst Chinese patients with lung cancer: feasibility study. J Med Internet Res. 2018;20:e11087.
56. Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLOS Med. 2018;15:e1002683.
57. Lambin P, Roelofs E, Reymen B, et al. ‘Rapid learning health care in oncology’: an approach towards decision support systems enabling customised radiotherapy. Radiother Oncol. 2013;109:159-64.
58. Ross JS, Waldstreicher J, Bamford S, et al. Overview and experience of the YODA Project with clinical trial data sharing after 5 years. Sci Data. 2018;5:180268.
59. London JW. Cancer research data-sharing networks. JCO Clin Cancer Inform. 2018;2:1-3.
60. ORIEN: Oncology Research Information Exchange Network. http://oriencancer.org/. Accessed December 3, 2018.
61. Academics & Hospitals. Flatiron Health. https://flatiron.com/academics/. Accessed December 3, 2018.
62. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018.
63. Chavan V, Penev L. The data paper: a mechanism to incentivize data publishing in biodiversity science. BMC Bioinformatics. 2011;12(suppl 15):S2.
64. Castelvecchi D. Can we open the black box of AI? Nat News. 2016;538:20.
65. Kuang C. Can A.I. be taught to explain itself? The New York Times. November 21, 2017. https://www.nytimes.com/2017/11/21/magazine/can-ai-be-taught-to-explain-itself.html. Accessed December 3, 2018.
66. Key Changes with the General Data Protection Regulation – EUGDPR. https://eugdpr.org/the-regulation/. Accessed December 3, 2018.
67. Towards trustable machine learning. Nat Biomed Eng. 2018;2:709-10.
68. Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719.
69. Zhang Q, Zhu SC. Visual interpretability for deep learning: a survey. February 2018. https://arxiv.org/abs/1802.00614. Accessed November 9, 2018.
70. Olah C, Satyanarayan A, Johnson I, et al. The building blocks of interpretability. Distill. https://distill.pub/2018/building-blocks/. Accessed February 14, 2019.
71. Educating the next generation of medical professionals with machine learning is essential. Science Daily. https://www.sciencedaily.com/releases/2018/09/180927083327.htm. Accessed November 12, 2018.
72. ACR-DSI. https://www.acrdsi.org/. Accessed December 3, 2018.
73. ASCO CancerLinQ. https://cancerlinq.org/. Accessed December 3, 2018.
74. American Society for Radiation Oncology (ASTRO). https://www.astro.org/Patient-Care-and-Research/Research. Accessed December 3, 2018.
75. National Institutes of Health. Big Data to Knowledge. NIH Common Fund. https://commonfund.nih.gov/bd2k. Accessed November 12, 2018.
76. Chen B, Ma L, Paik H, et al. Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat Commun. 2017;8:16022.
77. Lu Y, Yu Q, Gao Y, et al. Identification of metastatic lymph nodes in MR imaging with faster region-based convolutional neural networks. Cancer Res. 2018;78:5135-43.
78. Nikolov S, Blackwell S, Mendes R, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. September 2018. https://arxiv.org/abs/1809.04430. Accessed November 8, 2018.
79. Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. Med Image Comput Comput Assist Interv. 2013;16:254-61.
80. Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112:E6265-E6273.
81. Lao J, Chen Y, Li ZC, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353.
82. Yousefi S, Amrollahi F, Amgad M, et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci Rep. 2017;7:11707.
83. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
84. Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24:1248-59.
85. Feng Q, Dueva E, Cherkasov A, Ester M. PADME: a deep learning-based framework for drug-target interaction prediction. July 2018. http://arxiv.org/abs/1807.09741. Accessed February 14, 2019.
86. Preuer K, Lewis RPI, Hochreiter S, et al. DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics. 2018;34:1538-46.
87. Aliper A, Plis S, Artemov A, et al. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016;13:2524-30.

Late Hepatic Recurrence From Granulosa Cell Tumor: A Case Report
Granulosa cell tumors exhibit late recurrence and rare hepatic metastasis, emphasizing the need for lifelong surveillance in affected patients.