18-Gene Cluster Found in ER-Positive Breast Cancers
GAITHERSBURG, Maryland-Researchers have identified 18 genes that behave similarly to estrogen-receptor-alpha (ESR1) in patients with estrogen-receptor-positive (ER-positive) breast cancer. The cluster includes seven genes not previously associated with estrogen regulation or with breast cancer, Mihael H. Polymeropoulos, MD, head of pharmacogenetics, Novartis Pharmaceuticals Corporation, told ONI in an interview.
GAITHERSBURG, MarylandResearchers have identified 18 genes that behave similarly to estrogen-receptor-alpha (ESR1) in patients with estrogen-receptor-positive (ER-positive) breast cancer. The cluster includes seven genes not previously associated with estrogen regulation or with breast cancer, Mihael H. Polymeropoulos, MD, head of pharmacogenetics, Novartis Pharmaceuticals Corporation, told ONI in an interview.
Dr. Polymeropoulos and his colleagues described the cluster in an article in The Pharmacogenomics Journal (1:135-141, 2001). He said that some activity in the cluster might be coincidental to breast cancer, but all 18 genes should be investigated as potential biomarkers for predicting how individual patients will respond to endocrine therapies such as tamoxifen (Taxol) and letrozole (Femara). "This is the beginning of customizing the treatment of breast cancer," he told ONI.
ER-positive breast cancers are less likely to relapse and have better overall survival rates, he said, but about half do not respond to antiestrogens or estrogen-deprivation therapy. The hope is that supplementing ESR1 with additional markers will help physicians differentiate tumors by their genetic fingerprints and choose treatments accordingly.
The following seven genes had not previously been linked to estrogen stimulation or breast cancer: SCNNIA, SERPINA3, ASAH, LCN1, TGFBR3, GRIA2, and CYP2B. The other genes in the cluster are: CEACAM5, MGB1, LIV-1, PIP, MGP, TFF1, TFF3, HNF3A, HPN, XBP1, and AZGP1.
Dr. Polymeropoulos’ group found the genes via a DNA microarray, which can analyze gene expression at the level of RNA. In this process, he said, small glass slides containing minute amounts of thousands of known genes are matched against each biopsy sample. The gene "chips" on each slide can rapidly detect which of the thousands of genes are expressed in a human tissue sample.
In this study, the investigators examined gene expression for 7,000 genes in RNA taken from 53 breast tumors and from six other samples, including pooled normal breast tissue. That was as many genes as could be studied at the time, but Dr. Polymerop-oulos predicts that all 30,000 or more genes in the human genome will eventually be screened for overexpression in ER-positive breast cancer.
Twenty-one RNA samples in the study came from a randomized phase III clinical trial of letrozole vs tamoxifen that led to the FDA’s approval in January 2001 of letrozole tablets as a first-line hormonal treatment for advanced breast cancer in postmenopausal women.
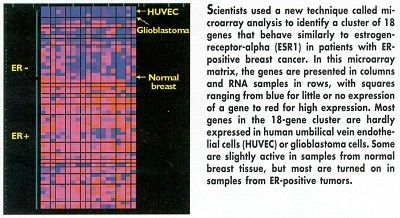
As most women in the trial were ER positive, the researchers added 30 samples of primary breast adenocarcinomas from a clinical trial in Sweden to raise the number of ER-negative tumors in the study. In addition, the researchers screened RNA from human umbilical vein endothelial cells, a glioblastoma cell line, and a pooled normal human breast sample.
First, the scientists identified a subset of 1,156 genes in which at least one of the RNA breast tumor samples had at least a 20-fold difference in expression when compared to normal breast tissue.
Then they whittled this subset down to 18 genes that were clearly co-clustered with ESR1. All of the 18 genes had distinctly different expression profiles from the normal breast sample and were not expressed at all or were expressed at very low levels in the endothelial and glioblastoma cell lines.
"Every gene in that class could potentially be examined as a new therapeutic target," Dr. Polymeropoulos said, inviting prospective studies into the 18 genes.
Another potential application, he said, would be a blood test measuring a protein expressed by one of the genes to identify women at higher risk of developing breast cancer and to gauge response to treatment. ESR1 does not circulate in the blood, but seven genes in the cluster encode secreted proteins: TFF1, TFF3, SERPINA3, PIP, TGFRB3, and AZGP1.
"Think about the PSA test in prostate cancerthis may be an analogy for breast cancer," Dr. Polymeropoulos suggested, describing TFF1 and TFF3 (trefoil factor 1 and 3) as the most likely candidates. TFF1 is already established as a biomarker for response to estrogen therapy. Estradiol is known to increase its expression, but tamoxifen can inhibit the effect.