Mutation-Derived Tumor Antigens: Novel Targets in Cancer Immunotherapy
Because of the abundance of promising preclinical and early-phase clinical data, mutation-derived tumor antigens an exciting new class of targets in cancer immunotherapy.
Oncology (Williston Park). 29(12):970-972, 974-975.

Figure 1. Identifying Mutation-Derived Tumor Antigens (MTAs)
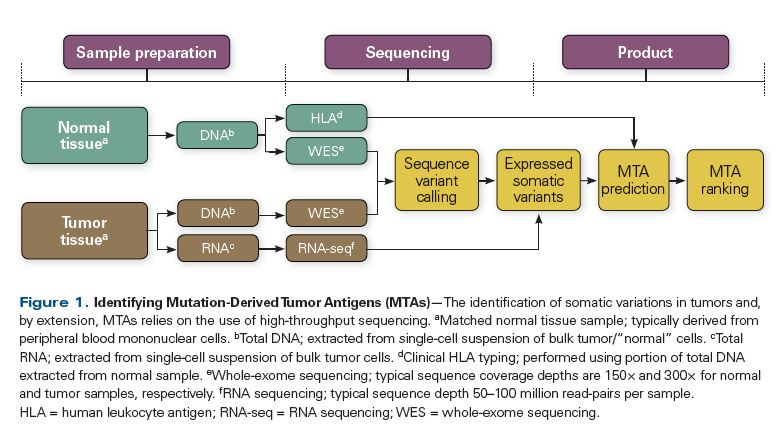
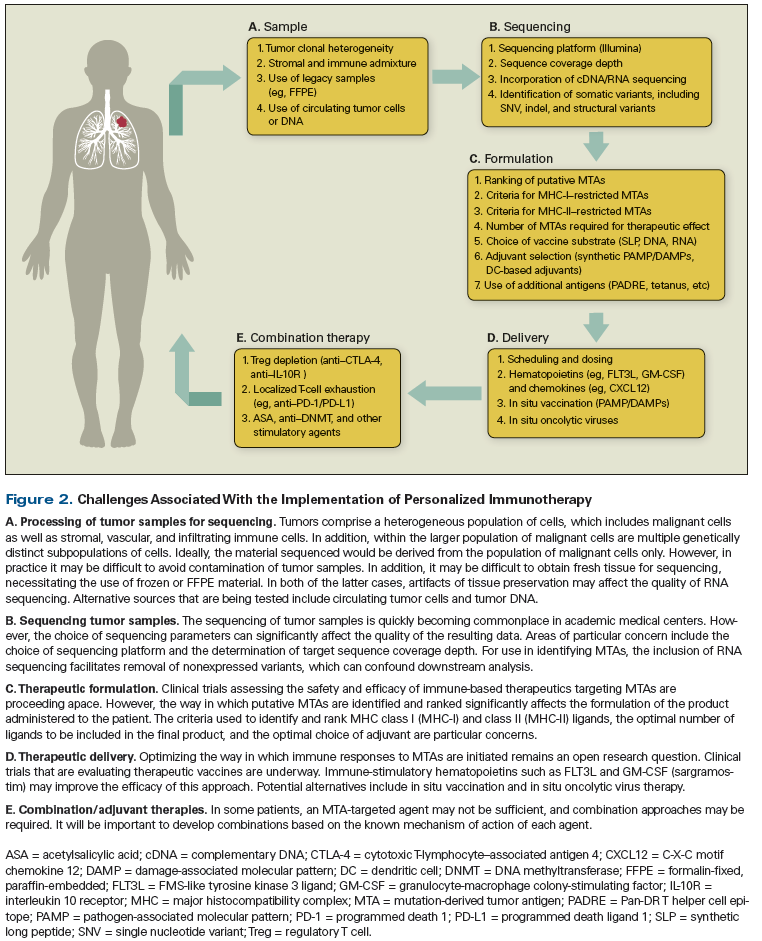
Figure 2. Challenges Associated With the Implementation of Personalized Immunotherapy
Introduction
The ability to leverage the adaptive immune system to treat human cancers has been a goal of immunologists and oncologists dating back to the pioneering experiments of William Coley. The ideal immunotherapy should mobilize the immune system to clear an existing population of malignant cells, without damaging adjacent healthy cells, and remain active in the event of recurrence. In practice, these goals have yet to be fully realized, in part because of the difficulties associated with identifying targets of the antitumor immune response. However, the introduction of highly effective immunotherapeutics-such as immune checkpoint blockade-coupled with advances in gene sequencing, has changed the face of the field entirely. Recent evidence indicates that mutations unique to a given individual’s tumor may be the long–sought-after targets of effective antitumor immunity and may hold the key to the design of the next generation of immune-based therapeutics.
Background
Descriptions of tumor antigens first appeared in the literature in the middle of the 20th century following observations of immune-mediated clearance of carcinogen-induced tumors in murine experimental models.[1-4] In the subsequent decades many tumor antigens have been identified, including developmental (carcinoembryonic antigen, WT1 [Wilms tumor 1]), lineage-specific (Melan-A/MART-1 [melanoma antigen recognized by T cells 1], prostatic acid phosphatase), and cancer/testis (MAGE-A [melanoma-associated antigens], NY-ESO-1, PRAME [preferentially expressed antigen in melanoma]) antigens, all of which are produced by genes overexpressed by malignant cells.[5] These antigens are easily identifiable and often shared among a cohort of subjects.[6,7] However, they are, by definition, “self” antigens and, like other “self” antigens, subject to immune tolerance.[8-11] Thus, the qualities that make these tumor antigens amenable to study may in turn limit their therapeutic utility.[12-15]
Recent advances in genetic sequencing technologies have facilitated the discovery of a new class of tumor antigen derived from somatic mutations, which we have termed mutation-derived tumor antigens (MTAs) but which are also known as neoantigens,or tumor-specific antigens.[16,17] MTAs are the product of nonsynonymous somatic variations randomly acquired by malignant cells as a result of deregulated progression through the cell cycle. Nonsynonymous somatic variations change protein sequence and function, facilitating the acquisition of traits associated with replicative advantage and tissue invasiveness.[18,19] Yet, when a nonsynonymous somatic variant is expressed and the variant sequence translated, the immune system may perceive the variant protein as “foreign” or “nonself” and respond to the cell as though it were infected with a pathogen.[20]
Notably, the likelihood of generating MTAs appears to be roughly proportional to the number of somatic mutations present within a given tumor.[21,22] However, the production of MTAs does not depend on the type of somatic variation. MTAs have arisen from substitutions, insertions, and deletions, as well as from larger structural rearrangements, such as duplications, inversions, and translocations. Nor does the production of MTAs depend on the oncologic significance of the instigating mutation. MTAs have arisen from both classic tumor suppressors and oncogenes, as well as from somatic mutations of unknown significance. Finally, the production of MTAs does not appear to depend on clinical characteristics of age, sex, and disease histology. MTAs have been identified in patients with multiple types of solid and hematologic malignancies.[23-37]
Evidence from both murine and human studies suggests that MTAs play a significant role in the endogenous adaptive immune response to tumors, as well as the induced adaptive immune response to tumors treated with cytotoxic T-lymphocyte–associated antigen 4 (CTLA-4) or programmed death 1 (PD-1) antagonists. Indeed, patients with melanoma or squamous-cell non–small-cell lung cancer who are treated with immune checkpoint blockade appear to stand a greater chance of clinical benefit as the burden of somatic mutations increases.[38-40] Furthermore, patients with tumors deficient in DNA mismatch repair, in which the number of somatic mutations can be multiple orders of magnitude greater than in tumors of comparable origin, also appear to fare significantly better when treated with immune checkpoint blockade.[41] However, prospective trials will be required to establish the utility of MTAs as predictive biomarkers. Given the considerable excitement brought about by these findings, as well as the ongoing development of therapeutic approaches that directly target MTAs, we will consider how these tumor antigens are identified, highlight deficiencies in current processes, and speculate as to how existing methods may be improved through further research.
Identification of MTAs
The identification of somatic variations in tumors and, by extension, MTAs relies on the use of high-throughput sequencing (Figure 1).[42-44] Typically, material from both the tumor and a matched normal sample are sequenced in tandem.[45,46] Due to economic and technical considerations, sequencing efforts are restricted to the known protein-coding regions of the genome (exome). However, the use of targeted sequencing panels is not advised because this severely limits the ability to identify novel MTAs.
Sequencing experiments designed to identify MTAs are performed at higher coverage depths than are typically encountered in the literature. This is true with regard to the sequencing of tumor material, as well as material derived from matched normal samples. Sequence coverage depth is described in terms of the mean number of reads successfully realigned to targeted loci. However, the process by which the exome is enriched prior to sequencing can result in significant nonuniformity in coverage depth. To achieve acceptable mean coverage across all target loci, a mean sequence coverage depth of 150× may be required for normal samples.[47] When tumor samples are sequenced, coverage depths are typically doubled (300×) to account for additional confounders, including intrinsic tumor heterogeneity and contamination of tumor samples with material from stromal and/or other infiltrating cell types. These and other sources of bias may be reduced even further through random sampling of multiple, distinct regions of the tumor or through sequencing of circulating tumor material (cells, DNA).
MTAs are produced when a nonsynonymous somatic variant is expressed and the variant sequence is translated into protein. The targeting of MTAs derived from expressed variants can lead to therapeutic responses in vivo in animal models, but the utility of MTAs derived from nonexpressed variants has not been established.[42-44,48] DNA sequencing can be used to measure sequence variation; however, it does not provide a measure of gene expression. Quantifying expression eliminates nonexpressed variants that can confound downstream analysis. In addition, expression analysis through RNA sequencing, as opposed to array-based methods, directly determines transcript sequence and structure, which mitigates the requirement for accurate annotation of the transcriptome. Unfortunately, because of both biologic and technical variables, it is difficult to provide a general recommendation with regard to target depth for RNA sequencing experiments. However, depths in excess of 50 to 100 million reads per sample can be required to unambiguously resolve the sequence of variant-containing transcripts.
The technical details of DNA and RNA sequence analysis are beyond the scope of this article. However, we would like to emphasize that the choice of parameters used in sequence analysis can introduce systematic error into the results of these analyses. To overcome these issues, it is advisable to perform all analyses with extensively vetted combinations of bioinformatics tools.[49] It may also be advisable to use many such vetted combinations and to consider only those results arrived at by consensus.[50,51]
The final step in identifying MTAs further refines the list of expressed nonsynonymous variants and eliminates those that are unlikely to be immunologically relevant. This procedure has been the subject of a number of recent reviews and will not be exhaustively detailed here. Fritsch et al, van Buuren et al, Rajasagi et al, and Gubin et al provide comprehensive overviews of the methodology.[52-55] Briefly, computational tools are used to assess the variant protein sequence and to determine whether it will be presented by host major histocompatibility complex (MHC) class I or II molecules-surface receptors required for the initiation of a cellular adaptive immune response. It has been demonstrated that putative MTAs that are predicted to bind to MHC molecules by this method can lead to therapeutic responses in vivo.[42-44] However, the criteria used to make this distinction were developed for the infectious disease setting, and additional studies will be required to formally establish criteria for use in identifying MTAs. Finally, there is an urgent need for new animal models that can be used to evaluate MTA-specific therapeutic approaches in a manner that can be translated into studies performed in humans.
Therapeutic Targeting of MTAs
Much remains to be determined regarding the specific dose, schedule, and route of delivery of MTA-targeting immune-based therapeutics and the relative role of CD4- vs CD8-mediated immunity (Figure 2).[48] At present, a number of early-phase clinical trials are assessing MTA-targeting vaccines, and the preliminary results are encouraging.[56] Tumor-lysate–, peptide-, RNA-, and dendritic cell–based vaccine substrates are being studied, but the optimal choice of vaccine adjuvant remains to be determined. Trials currently underway have largely employed certain synthetic toll-like receptor ligands, which have been studied in other vaccination projects. However, adjuvants that target other innate immune pattern recognition receptors, including C-type lectin (CLR), nucleotide-binding oligomerization domain (NOD), RIG-I–like (RLR), and stimulator of interferon genes (STING) receptor pathways, remain viable, unstudied alternatives. The use of stimulatory hematopoietins, such as FMS-like tyrosine kinase 3 ligand and granulocyte-macrophage colony-stimulating factor (sargramostim), should also be considered.[57-59]
Evidence suggests that indirect targeting of MTAs through the use of vaccine adjuvants or oncolytic viruses injected or infused in situ or directly into the body of existing tumors may represent yet another approach to the stimulation of an MTA-specific immune response.[60,61] Preclinical work suggests that in situ vaccination may be used as a bridge to formal MTA-targeted vaccination for patients with advanced primary or recurrent disease, thus broadening the range of patient groups who are eligible to receive precision immune-based therapeutics.
Finally, it will be important to consider the use of MTA-targeted agents in conjunction with CTLA-4 and PD-1/programmed death ligand 1 antagonists (see Figure 2). Although combination approaches have not been formally tested in humans, preliminary evidence suggests that MTA-targeted approaches can boost existing MTA-specific responses and also provoke MTA-specific responses de novo.[56] Thus, combination therapy may serve as a safe and effective means of overcoming resistance to immune checkpoint blockade.[61]
Conclusion
At present much remains to be determined regarding the efficacy of MTA-specific immune-based therapeutics, used individually or in combination with other therapeutics. Generating these data will require innovative prospective trial designs compatible with the degree of subject-to-subject variability inherent to this approach. However, because of the abundance of promising preclinical and early-phase clinical data, many in the field are initiating such trials, making the MTAs an exciting new class of targets in cancer immunotherapy.
Financial Disclosure:Dr. Bhardwaj is a cofounder of Check Point Sciences. The other authors have no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
References:
1. Foley EJ. Antigenic properties of methylcholanthrene-induced tumors in mice of the strain of origin. Cancer Res. 1953;13:835-7.
2. Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769-78.
3. Klein G, Sjogren HO, Klein E, Hellstrom KE. Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host. Cancer Res. 1960;20:1561-72.
4. Kripke ML. Antigenicity of murine skin tumors induced by ultraviolet light. J Natl Cancer Inst. 1974;53:1333-6.
5. Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135-46.
6. Vansteenkiste J, Zielinski M, Linder A, et al. Adjuvant MAGE-A3 immunotherapy in resected non-small-cell lung cancer: phase II randomized study results. J Clin Oncol. 2013;31:2396-403.
7. Kruit WH, Suciu S, Dreno B, et al. Selection of immunostimulant AS15 for active immunization with MAGE-A3 protein: results of a randomized phase II study of the European Organisation for Research and Treatment of Cancer Melanoma Group in Metastatic Melanoma. J Clin Oncol. 2013;31:2413-20.
8. Villaseñor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci USA. 2008;105:15854-9.
9. Giraud M, Yoshida H, Abramson J, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci USA. 2012;109:535-40.
10. Pinto S, Michel C, Schmidt-Glenewinkel H, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci USA. 2013;110:E3497-E3505.
11. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. 2014;14:377-91.
12. Theobald M, Biggs J, Dittmer D, et al. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci USA. 1995;92:11993-7.
13. Theobald M, Biggs J, Hernández J, et al. Tolerance to p53 by A2.1-restricted cytotoxic T lymphocytes. J Exp Med. 1997;185:833-41.
14. Morris GP, Ni PP, Allen PM. Alloreactivity is limited by the endogenous peptide repertoire. Proc Natl Acad Sci USA. 2011;108:3695-700.
15. Träger U, Sierro S, Djordjevic G, et al. The immune response to melanoma is limited by thymic selection of self-antigens. PLoS One. 2012;7:e35005.
16. Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747-52.
17. van Rooij N, van Buuren MM, Philips D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439-e442.
18. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70.
19. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74.
20. Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400-4.
21. Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743-50.
22. Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61.
23. Robbins PF, El-Gamil M, Li YF, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185-92.
24. Zorn E, Hercend T. A natural cytotoxic T cell response in a spontaneously regressing human melanoma targets a neoantigen resulting from a somatic point mutation. Eur J Immunol. 1999;29:592-601.
25. Chiari R, Foury F, De Plaen E, et al. Two antigens recognized by autologous cytolytic T lymphocytes on a melanoma result from a single point mutation in an essential housekeeping gene. Cancer Res. 1999;59:5785-92.
26. Dolstra H, Fredrix H, Maas F, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. 1999;189:301-8.
27. Torikai H, Akatsuka Y, Miyazaki M, et al. The human cathepsin H gene encodes two novel minor histocompatibility antigen epitopes restricted by HLA-A*3101 and -A*3303. Br J Haematol. 2006;134:406-16.
28. van Bergen CA, Kester MG, Jedema I, et al. Multiple myeloma-reactive T cells recognize an activation-induced minor histocompatibility antigen encoded by the ATP-dependent interferon-responsive (ADIR) gene. Blood. 2007;109:4089-96.
29. Hogan KT, Eisinger DP, Cupp SB, et al. The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res. 1998;58:5144-50.
30. Karanikas V, Colau D, Baurain JF, et al. High frequency of cytolytic T lymphocytes directed against a tumor-specific mutated antigen detectable with HLA tetramers in the blood of a lung carcinoma patient with long survival. Cancer Res. 2001;61:3718-24.
31. Echchakir H, Mami-Chouaib F, Vergnon I, et al. A point mutation in the alpha-actinin-4 gene generates an antigenic peptide recognized by autologous cytolytic T lymphocytes on a human lung carcinoma. Cancer Res. 2001;61:4078-83.
32. Linnebacher M, Gebert J, Rudy W, et al. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int J Cancer. 2001;93:6-11.
33. Ripberger E, Linnebacher M, Schwitalle Y, et al. Identification of an HLA-A0201-restricted CTL epitope generated by a tumor-specific frameshift mutation in a coding microsatellite of the OGT gene. J Clin Immunol. 2003;23:415-23.
34. Mandruzzato S, Brasseur F, Andry G, et al. A CASP-8 mutation recognized by cytolytic T lymphocytes on a human head and neck carcinoma. J Exp Med. 1997;186:785-93.
35. Guéguen M, Patard JJ, Gaugler B, et al. An antigen recognized by autologous CTLs on a human bladder carcinoma. J Immunol. 1998;160:6188-94.
36. Gaudin C, Kremer F, Angevin E, et al. A hsp70-2 mutation recognized by CTL on a human renal cell carcinoma. J Immunol. 1999;162:1730-8.
37. Wick DA, Webb JR, Nielsen JS, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20:1125-34.
38. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189-99.
39. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-8.
40. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA4 blockade in metastatic melanoma. Science. 2015;350:207-11.
41. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509-20.
42. Castle JC, Kreiter S, Diekmann J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081-91.
43. Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577-81.
44. Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572-6.
45. Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53.
46. Hiltemann S, Jenster G, Trapman J, et al. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res. 2015;25:1382-90.
47. Shigemizu D, Momozawa Y, Abe T, et al. Performance comparison of four commercial human whole-exome capture platforms. Sci Rep. 2015;5:12742.
48. Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692-6.
49. Xu H, DiCarlo J, Satya RV, et al. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics. 2014;15:244.
50. Goode DL, Hunter SM, Doyle MA, et al. A simple consensus approach improves somatic mutation prediction accuracy. Genome Med. 2013;5:90.
51. Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843-51.
52. Fritsch EF, Rajasagi M, Ott PA, et al. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522-9.
53. van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836.
54. Rajasagi M, Shukla SA, Fritsch EF, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453-62.
55. Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125:3413-21.
56. Carreno BM, Magrini V, Becker-Hapak M, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803-8.
57. Kreiter S, Diken M, Selmi A, et al. FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res. 2011;71:6132-42.
58. Anandasabapathy N, Feder R, Mollah S, et al. Classical Flt3L-dependent dendritic cells control immunity to protein vaccine. J Exp Med. 2014;211:1875-91.
59. Lim K, Hyun YM, Lambert-Emo K, et al. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science. 2015;349:aaa4352.
60. Salazar AM, Erlich RB, Mark A, et al. Therapeutic in situ autovaccination against solid cancers with intratumoral poly-ICLC: case report, hypothesis, and clinical trial. Cancer Immunol Res. 2014;2:720-4.
61. Woller N, Gürlevik E, Fleischmann-Mundt B, et al. Viral infection of tumors overcomes resistance to PD-1-immunotherapy by broadening neoantigenome-directed T-cell responses. Mol Ther. 2015;23:1630-40.
Late Hepatic Recurrence From Granulosa Cell Tumor: A Case Report
Granulosa cell tumors exhibit late recurrence and rare hepatic metastasis, emphasizing the need for lifelong surveillance in affected patients.