Clinical Applications of The Cancer Genome Atlas Project (TCGA) for Squamous Cell Lung Carcinoma
We summarize here key findings from the comprehensive analysis of squamous cell lung cancer by The Cancer Genome Atlas group and discuss the clinical implications of these findings.
Very little progress has been made in the treatment of patients with metastatic squamous cell lung cancer over the past 2 decades. Identification of novel molecular alterations for targeted therapies is necessary to improve outcomes. Advances in genomic technology have now made it possible to analyze the genomic landscape of tumor tissues comprehensively. We summarize here key findings from the comprehensive analysis of squamous cell lung cancer by The Cancer Genome Atlas group and discuss the clinical implications of these findings.
Introduction
Lung cancer remains the major cause of cancer-related death in the United States.[1] Approximately 85% of lung cancer patients are diagnosed with non–small cell lung cancer (NSCLC).[2,3] Adenocarcinoma and squamous cell lung carcinoma (SqCC) are the most commonly diagnosed histologic subtypes of NSCLC, and 20% to 30% of patients with NSCLC have SqCC histology.[2] Recent advances in the field of cancer biology and therapeutics have led to the successful development and approval of targeted therapies that have demonstrated substantial efficacy in clinical trials.[4-7] The use of targeted agents in lung cancer patients harboring epidermal growth factor receptor (EGFR) gene mutations or anaplastic lymphoma receptor tyrosine kinase (ALK) and c-ros oncogene 1, receptor tyrosine kinase (ROS1) gene rearrangements has been associated with dramatic response rates and improved progression-free survival (PFS).[4,7-11] Unfortunately, improved outcomes with these agents have largely been confined to patients with adenocarcinoma. At presentation, most patients with NSCLC are diagnosed with metastatic disease which is associated with a dismal 5-year survival rate of 2%.[12] The current standard of care for patients with metastatic NSCLC negative for EGFR or ALK alterations is empiric therapy with platinum doublets.[13,14] The choice of therapy for patients with NSCLC is mostly independent of the histology, with the exception of pemetrexed and the monoclonal antibody against the vascular endothelial growth factor (VEGF), bevacizumab, which are not indicated in SqCC because of decreased efficacy and excessive toxicity, respectively.[15-17] Outcomes with existing chemotherapeutic regimens are far from optimal in SqCC, however, and there is an urgent need to develop novel agents for use in these patients.
Cell of Origin and Pathologic Features
SqCCs can potentially develop from precursor lesions in the bronchial mucosa that are characterized by squamous metaplasia resulting from exposure to noxious stimuli, such as tobacco smoke. By a series of molecular insults that lead to progressive dysplasia, these lesions evolve into pre-neoplastic carcinoma in situ (CIS) and, eventually, neoplasia.[18] The cellular architecture of the bronchial mucosa is complex; varies along a proximal-to-distal axis; and is made up of several cell types, including basal, ciliated, secretory, Clara, and neuroepithelial cells (see Figure).[19,20] Basal cells are considered to be progenitor cells with a potential to give rise to ciliated, secretory, or Clara cells, and they play a key role in the regeneration of airway epithelium following injury.[19,21,22] It is possible that the basal cell serves as the cell of origin for SqCC of the lung.[23,24] These cells are characterized by the expression of tumor protein p63 (TP63), in particular the delta-N63 or p40 isoform,[25] which is a molecular marker that aids in the histologic diagnosis of SqCC.[26,27] This isoform is capable of inhibiting the actions of tumor protein p53 (TP53).[28]
Based on their histologic appearance, SqCCs are classified into papillary, clear cell, small cell, and basaloid subtypes according to the World Health Organization (WHO)/International Association for the Study of Lung Cancer (IASLC) classification.[29,30] Among these, the basaloid subtype is associated with minimal squamous differentiation and a poor prognosis.[31,32] The utility of this classification system for purposes of prognostication or guiding therapeutic decisions is otherwise limited.
FIGURE
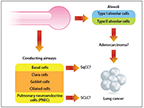
Schematic Representation of Major Cell Types in the Lung and Cell of Origin in Lung Cancer
Role of Cancer Genome Sequencing
Cancer is a disease of the cellular genome. Specific genetic alterations that confer “hallmark capabilities” to a cell facilitate emergence of the malignant phenotype.[33] These alterations are often acquired by cancer cells in a progressive manner, enabling them to silence tumor suppressor genes, gain limitless replicative potential, constitutively activate growth signaling, and develop metastatic potential in the process. Genes regulating these processes are also often implicated in organogenesis and cell differentiation. Cancer cells rely on the constitutive activity of certain oncogenic pathways for the sustenance of their neoplastic phenotype, in a phenomenon often referred to as “oncogene addiction.”[34] Some cancer cells are also known to exploit specific survival programs in the cellular genome that are associated with lineage development. Genes that mediate such differentiation programs are often referred to as “lineage-survival oncogenes.”[35] The advent of next-generation sequencing (NGS) technology has facilitated unbiased characterization of the cancer genome in unprecedented detail. A detailed understanding of the complex genetic pathways responsible for initiation and maintenance of malignant transformation may lead to identification of vulnerabilities in cancers, and in turn may enable identification of novel therapeutic targets.
TABLE 1
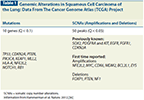
Genomic Alterations in Squamous Cell Carcinoma of the Lung: Data From The Cancer Genome Atlas (TCGA) Project
The complexity of the genetic aberrations driving SqCC were demonstrated clearly by recent preliminary findings reported from The Cancer Genome Atlas (TCGA) project; as a part of this project, 178 tumor samples and matched normal tissue samples were subjected to whole exome and transcriptome sequencing. Whole genome sequencing of 19 SqCC samples was also undertaken along with microRNA (miRNA) expression analysis of 159 samples. Averages of 228 non-silent mutations, 323 somatic copy number alterations (SCNAs), and 165 rearrangements were reported in this study. Overall, SCNAs involving 50 regions (Q < .05) and mutations in 10 genes (Q < .1) were considered to be significant.[36] The objective of this review is to briefly summarize the key findings reported by TCGA (Table 1) and discuss their clinical significance.
Key Pathway Alterations
Squamous cell differentiation pathway
Previous studies have reported frequent amplification of the chromosomal region 3q in SqCC of the lung, a finding that was confirmed by the TCGA.[36-38] Amplification of this segment has been considered to be crucial in the evolution of SqCC.[39] Genes of interest, such as SOX2, TP63, and phosphatidylinositol 3-kinase (PIK3CA), have been mapped to this chromosomal segment. The SOX2 gene plays a crucial role in guiding airway cell differentiation. During organogenesis, foregut cells destined to develop into the lung are characterized by SOX2 expression proximally.[20,40] Furthermore, SOX2 is also one of the factors capable of mediating the reprogramming of differentiated cells into pluripotent cells.[41] Basal cells in the epidermis of the skin and lung are known to express TP63, which plays a crucial role in maintaining epithelial self-renewal.[21,25,42-44] Overall, activation of the SOX2 and TP63 genes has been reported in 21% and 16% of TCGA samples, respectively. The Notch signaling pathway also plays an important role in cell differentiation. Activation of the Notch pathway guides the differentiation of cells in the airway.[22,45] Inactivating mutations in the Notch pathway can therefore block cell differentiation and possibly promote tumorigenesis.[43,46] Mutations in Notch genes have been reported in cutaneous and head and neck squamous cell carcinomas, a finding that further supports their role in SqCC oncogenesis.[46,47] NOTCH1 and NOTCH2 genes were inactivated in 8% and 5% of the samples analyzed by TCGA, respectively.[36] Taken together, these findings support the role for SOX2 as a “lineage-survival oncogene” and a possible tumor suppressor role for Notch signaling in SqCC of the lung.[43,46,48]
Oxidative stress response pathway
Expression of the nuclear factor erythroid 2–related factor 2 (Nrf2) aids cell survival in situations of oxidative stress by promoting expression of drug efflux pumps and enzymes mediating xenobiotic metabolism.[49,50] KEAP1 and CUL3 regulate cellular levels of Nrf2 by mediating its degradation. Therefore, overexpression of Nrf2 or inactivation of kelch-like ECH-associated protein 1 (KEAP1) and cullin-3 (CUL3) can confer on a cell a survival advantage in the setting of increased oxidative stress. Nrf2 activation in cancer cells has been found to confer protection from cytotoxic treatments and radiation therapy.[51,52] Activation of the Nrf2 pathway has been reported in 34% of the SqCC samples by TCGA.[36] Overexpression of Nrf2 or decreased expression of KEAP1 have been associated with poor outcomes in patients with SqCC.[53] Development of agents capable of inhibiting Nrf2 therefore carries the potential to improve outcomes in selected patients with SqCC.
Oncogene and tumor suppressor pathways
Loss of the tumor suppressor gene TP53 was reported in 90% of SqCC samples analyzed by TCGA.[36] Inactivation of another crucial tumor suppressor gene, CDKN2A, was also found in 72% of the samples by homozygous deletion, methylation, exon skipping, or inactivating mutations. Mutations in NF1, which functions as an inhibitor of Ras signaling, were reported in 11% of samples. A total of 47% of TCGA samples were considered to show activation of the phosphoinositide 3-kinase (PI3K) pathway, representing an important target for drug development.
Mutations in the oncogenes EGFR and Kirsten rat sarcoma viral oncogene homolog (KRAS) are infrequent in SqCC compared with their mutation frequency in adenocarcinoma.[54-56] EGFR amplification in 7% of the samples and mutations that were considered amenable for targeting with tyrosine kinase inhibitors (TKIs) in two samples were reported by TCGA.[36,57,58] Amplifications and nonrecurrent translocations involving the fibroblast growth factor receptor (FGFR) genes were also observed.[36,59] FGFR1, FGFR2, and FGFR3 were considered to be activated in 7%, 3%, and 2% of TCGA samples, respectively. Amplification of the chromosomal region 4q12, which contains genes for the tyrosine kinase receptors platelet-derived growth factor receptor alpha (PDGFRA) and Kit, has previously been reported in approximately 9% of SqCCs.[60] The PDGFRA gene was found amplified in nearly 4% of the cases analyzed by TCGA.[36,61]
Therapeutic Implications
Targetable alterations were considered to be present in 64% of the SqCC samples analyzed by TCGA.[36] Mutations or amplifications were reported among three families of targetable tyrosine kinases: ErbBs, Janus tyrosine kinases (JAKs), and FGFRs. At least one alteration involving PI3K/AKT, Ras, or tyrosine kinase pathways was observed in 69% of the samples. Of the mutations involving tyrosine kinases, 39% were present in the kinase domain. Overall, these findings suggest a potential role for agents targeting the PI3K, mammalian target of rapamycin (mTOR), EGFR, FGFR, and human rat sarcoma (RAS) pathways.
TABLE 2
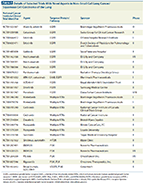
Details of Selected Trials With Novel Agents in Non–Small Cell Lung Cancer/Squamous Cell Carcinoma of the Lung
Tyrosine kinase inhibitors
Mutations that confer sensitivity to EGFR TKIs are rare in SqCC of the lung. Although modest responses have been reported with EGFR TKIs in patients harboring such mutations, these responses are usually inferior when compared to responses observed in adenocarcinomas.[62,63] Amplification of FGFR1 in SqCC has been associated with poor prognosis in patients with SqCC.[64] No clinical or demographic features were found to correlate with FGFR1 amplification in 226 SqCC patients analyzed by Heist and colleagues.[65] Agents targeting FGFRs, such as PD173074 and ponatinib, have showed inhibitory activity in lung cancer cell lines.[66,67] The multitargeted TKI cediranib, which targets VEGF receptor (VEGFR), FGFR, and PDGFR, was tested in combination with gemcitabine and carboplatin in patients with advanced NSCLC as first-line therapy.[68] This phase II trial failed to meet its primary endpoint of overall response rate of 40% but met its secondary endpoint of a progression-free survival (PFS) of 48% (95% confidence interval, 35%–62%) at 6 months (the protocol-specified threshold was 40%). The PDGFRs are also targeted by multitargeted TKIs, such as sunitinib, pazopanib, and nintedanib, in addition to cediranib. These agents are currently being tested in phase II and III trials in patients with advanced NSCLC (Table 2), and their efficacy in patients with SqCC remains to be established. Dasatinib is a TKI capable of targeting BCR-ABL, Src family tyrosine kinases, and ephrin receptor kinases.[69] Mutations in the DDR2 tyrosine kinase gene were reported in 3.2% (9 of 277) of primary SqCCs analyzed by Hammerman and colleagues, and in 1% of the cases analyzed by TCGA.[36,61,70] Sensitivity to dasatinib was reported in SqCC cell lines with DDR2 mutations.[70] Nevertheless, the use of dasatinib in patients with advanced NSCLC has only yielded modest outcomes to date. Of 34 patients with advanced NSCLC in a phase II trial with dasatinib, all of whom had adenocarcinoma, 4 showed stable disease for more than 6 months.[71] No biomarker, such as EGFR or KRAS mutational status or level of phosphorylated signal transducer and activator of transcription 3 (STAT3) or Src, seemed to correlate with PFS in this trial. Another phase II trial with dasatinib in patients with advanced SqCC of the lung (NCT01491633) was recently terminated due to safety concerns.[72] Identification of an appropriate biomarker capable of predicting response to dasatinib in patients with SqCC is therefore of critical importance.[73]
PI3K/AKT/mTOR pathway inhibitors
Buparlisib (BKM120), a class I PI3K inhibitor, was shown to be safe and well tolerated in a phase I study that recruited patients with advanced solid malignancies.[74] A synergistic effect between the mTOR inhibitor everolimus and BKM120 was observed in lung cancer cell lines and mouse xenograft models.[75] Trials with this agent in advanced NSCLC and SqCC are currently underway. To date, results with the protein kinase C and PI3K/AKT signaling inhibitor enzastaurin in patients with NSCLC have largely been disappointing.[76-78] A phase II trial with the AKT inhibitor MK2206 in combination with erlotinib is currently being evaluated in patients with advanced NSCLC. On the other hand, several trials to date have reported the use of mTOR inhibitors in patients with advanced NSCLC to be tolerable and safe.[79-83] However, the efficacy of these agents in patients with SqCC remains to be determined.
Cyclin-dependent kinase inhibitors
The cyclin-dependent kinase (CDK) inhibitor 2A (CDKN2A) gene is located on the chromosomal arm 9p21, and encodes for two proteins: p16INK4a and P14ARF.[84] These proteins function as tumor suppressors. CDK4 and CDK6 promote cell-cycle progression through phosphorylation and inactivation of RB1. CDK4 and CDK6 are, in turn, inhibited by p16INK4a, which can thereby induce a cell-cycle arrest. On the other hand, p14ARF interacts closely with the tumor suppressor TP53. As mentioned, CDKN2A inactivation was observed in 72% of TCGA samples, and CDK inhibitors have a potential role in the treatment of such tumors.[36]Flavopiridol is a multi-CDK inhibitor that has demonstrated antitumor activity in vitro and in human tumor xenografts.[85] Growth of rat lung adenocarcinoma cells lacking p16INK4a expression is inhibited by flavopiridol.[86] George and colleagues reported flavopiridol to be safe and tolerable when co-administered with carboplatin and paclitaxel in patients with NSCLC.[87] This drug is currently being tested in several clinical trials for hematopoetic and solid malignancies. Selective CDK inhibitors (CDK4/CDK6 inhibitors), such as PD-0332991 (palbociclib), NVP-LEE011, and LY2835219, have also shown encouraging results in preclinical studies.[88] Investigators on a phase III trial of palbociclib in breast cancer (PENELOPE-B) are now recruiting patients.[89]
TABLE 3

Molecular Subtypes of Squamous Cell Carcinoma of the Lung, and Possible Clinical Applications
Molecular subtypes of SqCC
Wilkerson and colleagues defined four molecular subtypes of SqCC based on data obtained from gene expression arrays: basal, classical, secretory, and primitive.[90] This subtype classification was reproducible by TCGA with a 94% level of agreement.[36] Each of these molecular subtypes is associated with characteristic pathway alterations and certain clinicopathologic features (Table 3). For instance, DNA repair–related gene sets were enriched in the primitive subtype, while tumors that belonged to the classical subtype were enriched for gene sets regulating xenobiotic metabolism, including the Nrf2 antioxidant pathway in these studies. Assigning patients to these subgroups therefore carries the potential to guide therapy and aid prognostication.
Future Directions
Although we have not made substantial progress in the treatment of patients with metastatic SqCC of the lung, identification of novel targets in this disease through TCGA work is certainly very encouraging and a step in the right direction. Over the next several months, TCGA will complete genomic analyses of 500 SqCCs of the lung. Well-designed clinical trials targeting key pathways in carefully selected patients are now desperately needed to translate these early promising findings to the clinic.
Financial Disclosure:Dr. Govindan has served as a consultant for Boehringer Ingelheim, Bristol-Myers Squibb, Covidien, Merck, and Pfizer. Drs. Devarakonda and Morgensztern have no significant financial interest or other relationship with the manufacturers of any products or providers of any service mentioned in this article.
References:
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30.
2. Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669-92.
3. Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the Surveillance, Epidemiologic, and End Results database. J Clin Oncol. 2006;24:4539-44.
4. Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693-703.
5. Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507-16.
6. O'Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994-1004.
7. Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735-42.
8. Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239-46.
9. Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121-8.
10. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947-57.
11. Shaw AT, Camidge DR, Engelman JA, et al. Clinical activity of crizotinib in advanced non-small cell lung cancer (NSCLC) harboring ROS1 gene rearrangement. J Clin Oncol. 2012;30:abstr 7508.
12. Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706-14.
13. Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92-8.
14. Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10:1236-71.
15. Paz-Ares L, De Marinis F, Dediu M, et al. PARAMOUNT: phase III study of maintenance pemetrexed (pem) plus best supportive care (BSC) versus placebo plus BSC immediately following induction treatment with pem plus cisplatin for advanced nonsquamous non-small cell lung cancer (NSCLC). J Clin Oncol. 2011;29 (suppl): abstr CRA7510.
16. Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543-51.
17. Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542-50.
18. Wistuba II, Gazdar AF. Lung cancer preneoplasia. Annu Rev Pathol. 2006;1:331-48.
19. Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493-512.
20. Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8-23.
21. Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 2010;3:545-56.
22. Rock JR, Gao X, Xue Y, et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639-48.
23. Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? Am J Respir Crit Care Med. 2007;175:547-53.
24. Hanna JM, Onaitis MW. Cell of origin of lung cancer. J Carcinog. 2013;12:6.
25. Nobre AR, Albergaria A, Schmitt F. p40: a p63 isoform useful for lung cancer diagnosis-a review of the physiological and pathological role of p63. Acta Cytol. 2013;57:1-8.
26. Bishop JA, Teruya-Feldstein J, Westra WH, et al. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol. 2012;25:405-15.
27. Pelosi G, Fabbri A, Bianchi F, et al. ÎNp63 (p40) and thyroid transcription factor-1 immunoreactivity on small biopsies or cellblocks for typing non-small cell lung cancer: a novel two-hit, sparing-material approach. J Thorac Oncol. 2012;7:281-90.
28. Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199-207.
29. Travis WD, Colby TV, Corrin B, et al. World Health Organization international histological classification of tumours. histological typing of lung and pleural tumours, 3rd ed. New York: Springer; 1999.
30. Brambilla E, Travis WD, Colby TV, et al. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059-68.
31. Brambilla E, Moro D, Veale D, et al. Basal cell (basaloid) carcinoma of the lung: a new morphologic and phenotypic entity with separate prognostic significance. Hum Pathol. 1992;23:993-1003.
32. Moro-Sibilot D, Lantuejoul S, Diab S, et al. Lung carcinomas with a basaloid pattern: a study of 90 cases focusing on their poor prognosis. Eur Respir J. 2008;31:854-9.
33. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74.
34. Weinstein IB. Cancer. Addiction to oncogenes-the Achilles heal of cancer. Science. 2002;297:63-4.
35. Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593-602.
36. Hammerman PS, Hayes DN, Wilkerson MD, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519-25.
37. Kang JU, Koo SH, Kwon KC, et al. Identification of novel candidate target genes, including EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of the lung. BMC Cancer. 2009;9:237.
38. Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J Thorac Oncol. 2008;3:212-5.
39. McCaughan F, Pole JC, Bankier AT, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med. 2010;182:83-91.
40. Whitsett JA, Haitchi HM, Maeda Y. Intersections between pulmonary development and disease. Am J Respir Crit Care Med. 2011;184:401-6.
41. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-72.â©
42. Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol. 1999;113:1099-105.
43. Nguyen BC, Lefort K, Mandinova A, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028-42.
44. Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714-8.
45. Collins BJ, Kleeberger W, Ball DW. Notch in lung development and lung cancer. Semin Cancer Biol. 2004;14:357-64.
46. Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108:17761-6.
47. Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-60.
48. Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238-42.
49. Ishii T, Itoh K, Takahashi S, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023-9.
50. Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313-22.
51. Wang XJ, Sun Z, Villeneuve NF, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235-43.
52. Singh A, Bodas M, Wakabayashi N, et al. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010;13:1627-37.
53. Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743-53.
54. Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945-50.
55. Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958-67.
56. An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7:e40109.
57. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129-39.
58. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497-500.
59. Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636-47.
60. Ramos AH, Dutt A, Mermel C, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009;8:2042-50.
61. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4.
62. Duan JC, An TT, Wu MN, et al. [Correlation between the efficacy of epidermal growth factor receptor tyrosine kinase inhibitors and EGFR mutations in advanced squamous cell lung cancer]. Zhonghua Jie He He Hu Xi Za Zhi. 2012;35:323-8.
63. Shukuya T, Takahashi T, Kaira R, et al. Efficacy of gefitinib for non-adenocarcinoma non-small-cell lung cancer patients harboring epidermal growth factor receptor mutations: a pooled analysis of published reports. Cancer Sci. 2011;102:1032-7.
64. Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731-7.
65. Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775-80.
66. Ren M, Hong M, Liu G, et al. Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep. 2013;29:2181-90.
67. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93.
68. Dy GK, Mandrekar SJ, Nelson GD, et al. A randomized phase II study of gemcitabine and carboplatin with or without cediranib as first-line therapy in advanced non-small-cell lung cancer: North Central Cancer Treatment Group Study N0528. J Thorac Oncol. 2013;8:79-88.
69. Lindauer M, Hochhaus A. Dasatinib: recent results Cancer Res. 2010;184:83-102.
70. Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1:78-89.
71. Johnson FM, Bekele BN, Feng L, et al. Phase II study of dasatinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:4609-15.
72. Dasatinib in advanced squamous cell lung cancer. NCI clinical trial ID number NCT01491633. Available from: http://www.clinicaltrials.gov/ct2/show/NCT01491633?term=NCT01491633&rank=1.
73. Kruser TJ, Traynor AM, Wheeler DL. The use of single-agent dasatinib in molecularly unselected non-small-cell lung cancer patients. Expert Opin Investig Drugs. 2011;20:305-7.
74. Bendell JC, Rodon J, Burris HA, et al. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282-90.
75. Ren H, Chen M, Yue P, et al. The combination of RAD001 and NVP-BKM120 synergistically inhibits the growth of lung cancer in vitro and in vivo. Cancer Lett. 2012;325:139-46.
76. Socinski MA, Raju RN, Stinchcombe T, et al. Randomized, phase II trial of pemetrexed and carboplatin with or without enzastaurin versus docetaxel and carboplatin as first-line treatment of patients with stage IIIB/IV non-small cell lung cancer. J Thorac Oncol. 2010;5:1963-9.
77. Vansteenkiste J, Ramlau R, von Pawel J, et al. A phase II randomized study of cisplatin-pemetrexed plus either enzastaurin or placebo in chemonaive patients with advanced non-small cell lung cancer. Oncology. 2012;82:25-9.
78. Casey EM, Harb W, Bradford D, et al. Randomized, double-blinded, multicenter, phase II study of pemetrexed, carboplatin, and bevacizumab with enzastaurin or placebo in chemonaive patients with stage IIIB/IV non-small cell lung cancer: Hoosier Oncology Group LUN06-116. J Thorac Oncol. 2010;5:1815-20.
79. Papadimitrakopoulou VA, Soria JC, Jappe A, et al. Everolimus and erlotinib as second- or third-line therapy in patients with advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7:1594-601.
80. Ramalingam SS, Owonikoko TK, Behera M, et al. Phase II study of docetaxel in combination with everolimus for second- or third-line therapy of advanced non-small-cell lung cancer. J Thorac Oncol. 2013;8:369-72.
81. Waqar SN, Gopalan PK, Williams K, et al. A phase I trial of sunitinib and rapamycin in patients with advanced non-small cell lung cancer. Chemotherapy. 2013;59:8-13.
82. Reungwetwattana T, Molina JR, Mandrekar SJ, et al. Brief report: a phase II "window-of-opportunity" frontline study of the MTOR inhibitor, temsirolimus given as a single agent in patients with advanced NSCLC, an NCCTG study. J Thorac Oncol. 2012;7:919-22.
83. Milton DT, Riely GJ, Azzoli CG, et al. Phase 1 trial of everolimus and gefitinib in patients with advanced non-small-cell lung cancer. Cancer. 2007;110:599-605.
84. Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001-14.
85. Kelland LR. Flavopiridol, the first cyclin-dependent kinase inhibitor to enter the clinic: current status. Expert Opin Investig Drugs. 2000;9:2903-11.
86. Honoki K, Yoshitani K, Tsujiuchi T, et al. Growth inhibition and induction of apoptosis by flavopiridol in rat lung adenocarcinoma, osteosarcoma and malignant fibrous histiocytoma cell lines. Oncol Rep. 2004;11:1025-30.
87. George S, Kasimis BS, Cogswell J, et al. Phase I study of flavopiridol in combination with paclitaxel and carboplatin in patients with non-small-cell lung cancer. Clin Lung Cancer. 2008;9:160-5.
88. Roberts PJ, Bisi JE, Strum JC, et al. Multiple roles of cyclin-dependent kinase 4/6 inhibitors in cancer therapy. J Natl Cancer Inst. 2012;104:476-87.
89. A Study of palbociclib in addition to standard endocrine treatment in hormone receptor positive HER2 normal patients with residual disease after neoadjuvant chemotherapy and surgery (PENELOPE-B). Available from: http://www.clinicaltrials.gov/ct2/show/NCT01864746?term=penelopeb&rank=1.
90. Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864-75.
91. Sutherland K, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397-403.
92. ClinicalTrials.gov. Drug information available from: http://www.clinicaltrials.gov/ct2/home.
93. Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci USA. 2007;104:19589-94.
Late Hepatic Recurrence From Granulosa Cell Tumor: A Case Report
Granulosa cell tumors exhibit late recurrence and rare hepatic metastasis, emphasizing the need for lifelong surveillance in affected patients.