New Biomarkers in Prostate Cancer
In this article, we review recent advances in the discovery of prostate cancer biomarkers, their integration into clinical practice, and implications for improving clinical management of the disease.
Figure 1: Biomarkers That Are Relevant to Screening for Prostate Cancer
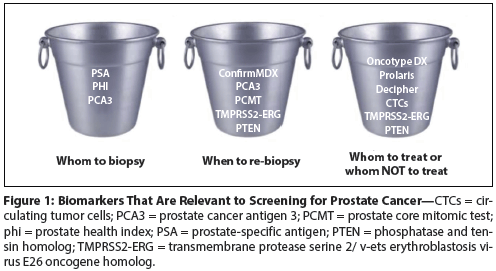
Figure 2: The Prostate Health Index (phi) Score Correlates With the Percentage of Positive Prostate Biopsies
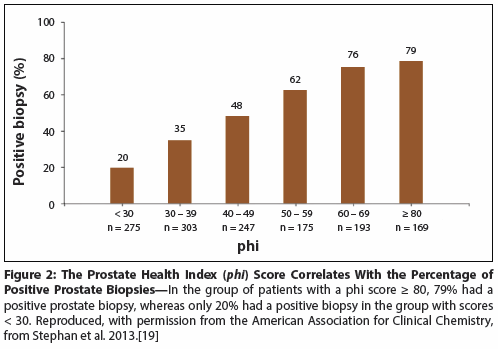
Figure 3: Percentage of Men With Positive Biopsy by Prostate Cancer Antigen 3 (PCA3) Score
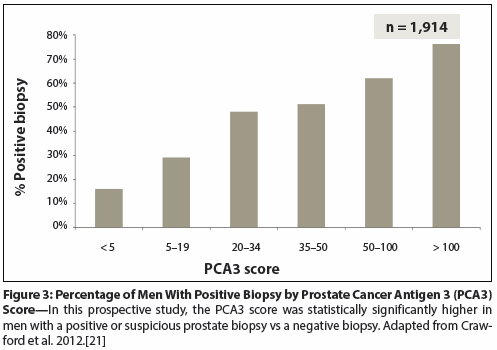
Figure 4: The Methylation Analysis to Locate Occult Cancer (MATLOC) Validation Study
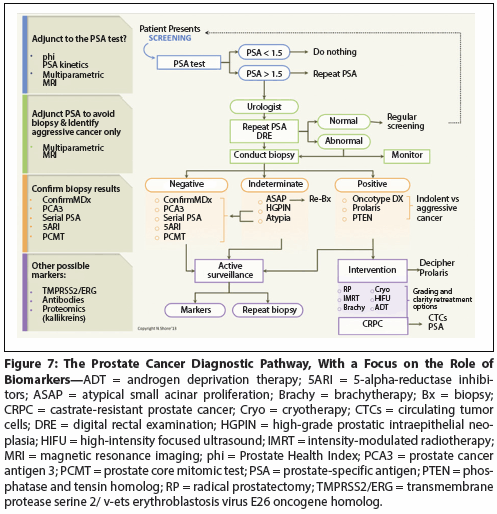
Figure 5: Assessing Individual Tumor Biology Using the Genomic Prostate Score (GPS)
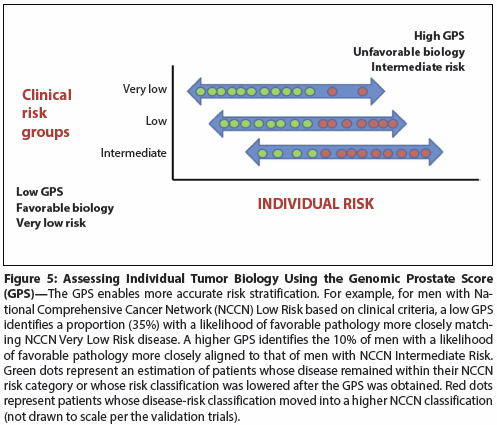
Figure 6: Risk Stratification of Prostate Cancer Patients Using Decipher, a Prognostic Genomic Marker Test
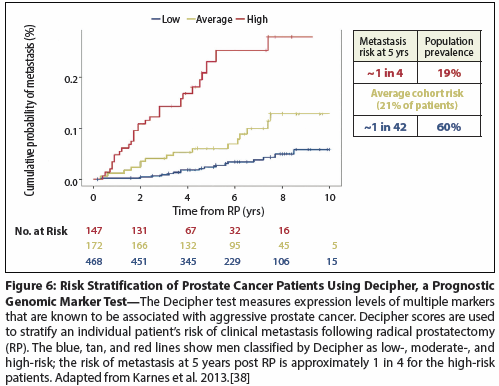
Figure 7: The Prostate Cancer Diagnostic Pathway, With a Focus on the Role of Biomarkers
Prostate cancer (PCa) is the most commonly diagnosed male cancer in the United States. The prostate-specific antigen (PSA) biomarker has been widely used to screen men for prostate cancer. Challenges of PSA cancer- specific sensitivity and specificity exist; fortunately, a new generation of PCa biomarkers is emerging, consisting of serum-, urine-, and tissue-based assays that may supplement PSA testing, or replace it over time. The identification and development of these biomarkers have been facilitated, in large part, by new genomic technologies that have enabled an additional interpretation of the individual patient’s tumor biology. Several biomarkers with specific indications for disease diagnosis, prediction, prognosis, and therapeutic response are now commercially available. Furthermore, genomic assays may now stratify the risk of aggressive PCa at the time of diagnosis. In this article, we review recent advances in the discovery of PCa biomarkers, their integration into clinical practice, and implications for improving clinical management of the disease.
Introduction
In the United States (US), approximately 240,000 men are diagnosed annually with prostate cancer (PCa).[1] Although effective treatment options are available for clinically localized PCa, the potential burdensome morbidities and attendant healthcare costs[2] from overdiagnosis and overtreatment have escalated the discussion and controversy regarding appropriate screening, diagnosis, and optimal management of PCa.[3,4]
Although the lifetime risk of developing PCa is approximately 1 in 6 (~16%),[5,6] the risk of dying from the disease is only ~2%.[6,7] The discrepancy between PCa incidence and lethality has led to widespread scrutiny of prostate cancer patient management, particularly for low-grade, low-stage (indolent) disease. The vast majority of men diagnosed with clinically localized PCa are treated with interventional therapies despite studies demonstrating that even without treatment, PCa-specific mortality is low.[8,9] Several factors may influence overtreatment; however, a very significant reason is that current clinical parameters are limited in their ability to discriminate between aggressive and indolent forms of the disease in a significant number of men.[10] Thus, clinicians and patients may lack sufficient confidence to comfortably select and maintain an active surveillance (active monitoring) strategy, for fear of missing a more aggressive phenotypic variant of the disease.
Given that PCa is both a biologically and clinically heterogeneous disease that develops amidst diverse genetic and epigenetic changes,[11,12] identification of disease-specific molecular biomarkers is a rational approach to addressing current clinical challenges of whom to biopsy, whom to offer certain interventional therapies, and in whom to alter therapeutic strategies.
Currently, biomarker research is focused on serum-, urine-, and tissue- based markers. Assays involving these biomarkers are being assessed to help patients avoid unnecessary biopsies; to reduce the use of interventional strategies, when clinically appropriate; and to enhance the risk stratification of organ-confined tumors. Markers are being developed to assist with the monitoring of progression during “watchful waiting,” to detect micrometastatic disease (below the limit of detection for imaging), and to better evaluate therapeutic responses to ongoing therapies.
PCa biomarkers can be broadly categorized into three main buckets depending on their role/utility in PCa management (Figure 1): biomarkers that assist clinicians in determining (1) whom to biopsy, (2) when to re-biopsy, and (3) whom to offer therapy.
Biomarkers That Help Clinicians Determine Whom to Biopsy
Prostate-specific antigen (PSA)
After its approval by the US Food and Drug Administration (FDA) in 1986, the PSA test revolutionized the PCa screening and diagnosis landscape. It should be remembered that PSA testing is approved for early detection along with digital rectal examination (DRE) in men over age 50.[13,14] In the US, approximately 19 million men are screened annually with PSA testing, resulting in more than 1.3 million biopsy procedures and 240,890 new diagnoses of PCa.[1]
Nonetheless, there are inherent limitations to using the PSA test for PCa screening. First, the test may give false-positive or false-negative results. Most men with an elevated PSA level (above 4.0 ng/mL)[15] are not found to have PCa; only about 25% of men who undergo a prostate biopsy due to an elevated PSA level actually have PCa. Conversely, a negative result may give false assurances that PCa is not detected, when in fact a cancer may exist. Also, early detection of PCa may not reduce a man’s chance of dying from the disease.[15]
The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial was a large population-based
randomized study designed and sponsored by the National Cancer Institute (NCI) to determine the effects of screening on cancer-related mortality and secondary endpoints in men and women aged 55 to 74. After 13 years of follow-up in the PLCO trial, there was no evidence of a mortality benefit for organized annual PCa screening vs opportunistic screening, which forms part of usual care, and there was no apparent interaction with age, baseline comorbidity, or pretrial PSA testing.[16] However, Crawford and colleagues, in assessing men with no comorbidities in the PLCO study, determined that screening did result in a survival benefit.[17]
Notably, however, 11-year follow-up results from the European Randomized Study of Screening for Prostate Cancer demonstrated that screening does significantly reduce death from PCa.[18] A potential reason for these different results is that in the US-based PLCO Cancer Screening Trial, at least 44% of participants in the control arm had already undergone PSA testing prior to being randomized into the study.[16]
To improve the sensitivity and specificity of serum PSA testing, several PSA derivatives and isoforms (eg, PSA isoforms and PSA density) may be used. PSA density may be useful in determining the presence of PCa.
Prostate health index (phi)
The phi (phi = [−2] proPSA/fPSA × PSA1/2; proPSA is a PSA subtype and fPSA is free PSA) has been developed as an additional diagnostic biomarker in men with a serum PSA level of 2–10 ng/mL. Previous studies have shown that elevated proPSA/fPSA ratios are associated with PCa. In a recent prospective cohort of men enrolled into active surveillance for PCa, higher serum and tissue levels of proPSA at diagnosis were associated with the need for subsequent treatment (Figure 2).[19]
Prostate cancer antigen 3 (PCA3)
PCA3 is a noncoding messenger RNA that has been shown to be elevated in > 90% of men with PCa, but not significantly elevated in normal prostatic glands or in benign prostatic hypertrophy. PCA3 is unique in that it can be measured in urine and adds to the diagnostic information obtained from the PSA test, with higher area under the curve (AUC) values of 0.66 to 0.72, compared with 0.54 to 0.63 for serum PSA alone.[20] PCA3 complements the PSA test in men undergoing initial biopsy.[21] In 2012, PCA3 was approved by the FDA as a diagnostic test for PCa in the setting of a previous negative prostate biopsy. PCA3 is also considered to be helpful in deciding when to re-biopsy and in the follow-up of patients under active surveillance (Figure 3).[21] The mean PCA3 score was statistically significantly higher in men with a positive biopsy, or those with atypical small acinar proliferation (ASAP) and/or high-grade prostatic intraepithelial neoplasia (HGPIN), compared with men who had a negative biopsy.[21]
Biomarkers That Help Clinicians Determine When to Re-biopsy
Up to 25% of men who undergo a prostate biopsy are misdiagnosed with a false-negative result. Unnecessary prostate biopsies subject many men to undue risk. Surveillance, Epidemiology, and End Results (SEER) Program data report that 6.9% of men are hospitalized in the 30 days following a prostate biopsy. Associated complications include bleeding, infection, sepsis/bacteremia, endocarditis, urinary symptoms/retention, and sexual dysfunction. Tests are needed to help determine who needs to be re-biopsied.
ConfirmMDx
ConfirmMDx is an epigenetic assay to help distinguish patients who have a true-negative biopsy from those who may have occult cancer. It detects an epigenetic field effect with the “cancerization” process at the DNA level. This field effect around the cancer lesion can be present despite the normal appearance of cells. Detection of field effects extends the coverage of the biopsy, helping to rule in, or rule out, occult cancers. It provides actionable information to help men without PCa avoid unnecessary repeat biopsies, with their inherent risk, and to identify men who require repeat biopsies and potential treatment.[22,23]
The MATLOC (Methylation Analysis To Locate Occult Cancer) validation study demonstrated the actionable and cost-effective nature of the ConfirmMDx test (also see the Cost vs Value section of this article). MATLOC is a blinded multicenter study of an epigenetic test for prostate cancer evaluating 483 men with initial negative biopsy followed by negative or positive biopsy. The real power of this test is in its negative predictive value of > 90% and its cost-effectiveness, as demonstrated in studies done on 1 million lives (Figure 4).[24,25]
Prostate Core Mitomic Test (PCMT)
The PCMT identifies a large-scale depletion in mitochondrial DNA that indicates cellular change associated with undiagnosed prostate cancer. It detects the presence of malignant cells in normal-appearing prostate tissue across an extended area.[26]
TMPRSS2-ERG
TMPRSS2-ERG is a fusion between the transmembrane protease serine 2 (TMPRSS2) gene and the v-ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) gene. This gene fusion is the predominant variant in approximately 40% to 80% of PCa. Quantitative levels of urine TMPRSS2-ERG appear to be associated with clinically significant PCa based on Epstein criteria, which stratify disease aggressiveness using PSA density and characteristics of the patient’s biopsy (Gleason score [GS], the percentage of tumor vs normal prostate tissue observed, and number of cores with tumor).[27]
Combining biomarker assays may improve predictive accuracy compared with the use of individual markers.[28] Assessment of post-DRE urine TMPRSS2-ERG, in combination with urine PCA3, enhanced the utility of serum PSA level for predicting PCa risk and clinically relevant cancer on subsequent biopsy.[29]
The PTEN gene
Dysregulation of PTEN, a tumor suppressor gene involved in cell cycle regulation, is consistently associated with poor prognosis in PCa. A preponderance of evidence shows that deletion of PTEN is associated with higher Gleason grade, risk of progression, and recurrence after therapy. Additionally, it is associated with advanced localized or metastatic disease and death.[30]
The PTEN assay is a prognostic fluorescence in situ hybridization (FISH) test typically ordered in conjunction with prostate biopsy tests to indicate partial (hemizygous) or complete (homozygous) deletions in the gene. For patients with a cancer diagnosis (for example, low-intermediate Gleason scores of 6/7), the PTEN assay may help determine the rate of progression and subsequent appropriate therapy. For patients with HGPIN or an atypical diagnosis, it is an effective screening tool that allows clinicians to distinguish nonaggressive HGPIN or an atypical diagnosis from men at higher risk of PCa.[31]
ProMark
ProMark is a prognostic biopsy-based PCa test. It uses immunofluorescent imaging analysis to quantify biomarker expression and classify patients’ tumors. A clinical validation study demonstrated ProMark can differentiate indolent from aggressive disease, based on data from standard formalin-fixed, paraffin-embedded tissue.[32]
Prostate Biomarkers That Help Clinicians Determine Whom to Treat or Not Treat
Given frequent uncertainty about the preferred course of treatment in many cases of early PCa, accurate prognostic markers that supplement standard clinicopathologic parameters are needed. Personalized risk assessment allows more informed treatment decision-making, given the variety of options available for management of early-stage disease. A number of biomarkers are available to guide clinicians through these challenging clinical situations.
Oncotype DX
Despite the fact that PCa remains a common cause of cancer death worldwide, many men will have indolent disease that will not threaten their health during their natural life span, and overtreatment of low-risk disease with radical therapy leads to significant morbidity and compromised quality of life.
Because of early detection efforts, the vast majority of men with PCa are diagnosed with clinically localized disease, yet are treated aggressively. However, studies indicate that even without treatment, PCa-specific mortality remains relatively low. This aggressive treatment is due primarily to the uncertainty around the malignant potential of many cancers. Improved prognostic tests that afford better stratification of patients based on their disease-specific risk of mortality and likelihood of progression allow more indolent disease to be treated conservatively and more aggressive disease to be treated more appropriately.
In men who have very low–, low-, and low-intermediate–risk PCa, Oncotype DX has been prospectively validated as a biopsy-based predictor of adverse pathology. The Genomic Prostate Score (GPS) adds independent predictive information beyond standard clinical and pathologic measures (Figure 5). The GPS assesses underlying biology from very small biopsy tumor volumes, addressing issues of tumor heterogeneity and biopsy under-sampling to more accurately predict disease aggressiveness. Incorporation of the GPS enables more accurate identification of a larger population of patients who can more confidently choose active surveillance as an initial management strategy. This test has been validated from biopsy material and adds to other clinical parameters such as National Comprehensive Cancer Network (NCCN) scores.[33,34]
Prolaris
Although overtreatment has garnered the majority of recent attention, under-treatment and resulting mortality of men who harbor more aggressive cancer can also remain a significant problem. These men may benefit from multimodality therapy, such as adjuvant radiation therapy, which may result in decreased disease-specific mortality. A key distinguishing factor between cancerous and noncancerous cells is the increase in cell cycle progression (CCP) gene mutations.[35] Prolaris is a prognostic genomic assay that assesses the CCP gene signature that has been validated in multiple cohorts and provides a risk assessment of PCa-specific progression and disease-specific mortality when combined with standard clinicopathologic parameters. Prolaris has been validated to provide personalized identification of both low-risk patients who can be managed with conservative options and high-risk patients who may benefit from earlier definitive treatment.[35]
Decipher
Approximately 50% of men with PCa who receive radiotherapy after RP derive no benefit from this intervention.[36] In men at elevated risk of recurrence following RP, only ~6% are found to have risk factors suggesting that they will develop biochemical progression and metastatic disease after 5 years.[37] Decipher is a genomic assay that assesses the risk of disease progression after radical prostatectomy. This assay has been shown to be independently prognostic of PCa death in a high-risk surgical cohort. In a validation study, over 70% of high-risk patients had low genomic classifier (GC) scores and good prognosis, whereas patients with high GC scores had a cumulative incidence of metastasis > 25%.[37] This assay may better enable application of directed, multimodal therapy for individual patients with high-risk PCa. The Decipher test is used to individualize the management of high-risk patients (differentiating men who may have a treatment benefit from those who may not) and may have a financial impact on use of radiation therapy or hormonal therapy (Figure 6).[38]
Biomarkers Used to Assess the Therapeutic Response
PSA does not always accurately reflect response to therapy for castration-resistant PCa (CRPC). Newer therapies, such as sipuleucel-T and radium-223, may improve survival without decreasing PSA levels. For cytotoxic or hormonal therapies, PSA responses are not always indicative of clinical response. Radiographic progression and symptomatology are still key parameters for consideration of changing antineoplastic therapy. Discordance between PSA kinetics and clinical response and progression of disease has been regularly observed. The need for biomarkers beyond PSA to predict response to treatment is well recognized. Other helpful serologic tests include hemoglobin, alkaline phosphatase, lactate dehydrogenase (LDH), and others. However, there is an unmet need for novel biomarkers to better assist clinical evaluation of therapeutic response in metastatic CRPC.
Circulating tumor cells (CTCs)
The CTC assay is intended for enumeration of CTCs (CD45−, epithelial cell adhesion molecule [EpCAM]+, and cytokeratins) in whole blood, which can be a biomarker for a therapeutic response to antineoplastic regimens. Increased levels of CTCs in the blood of CRPC patients can predict worse outcomes. Evaluation of individual CTCs has allowed further prognostication of PCa.[39] CTCs may be useful for predicting treatment response and survival outcomes associated with cytotoxic and hormonal therapies, but approximately 50% of patients have undetectable CTC levels based on current detection methods.[40] CTC detection techniques with improved sensitivity are under investigation.
Implications for Clinical Practice Treatment Guidelines
Despite extensive research efforts, few PCa biomarkers have been successfully validated and integrated into clinical practice (Figure 7). The serum-based PSA test therefore remains an important biomarker for PCa detection and follow-up. One of the important lessons learned from the popularization of PSA as a screening test is that biomarker development requires a priori deliberation of the intended role of a particular biomarker. The recent decision by the United States Preventive Services Task Force (USPSTF) to recommend against widespread PSA-based screening for PCa stems from findings that, even without treatment, PCa-specific mortality remains low. The Task Force concluded that the prevalence of overtreatment harms more patients than it benefits. The American Urological Association (AUA) initially opposed the recommendation but has since modified its position to partially align with that of the Task Force.
The potential complications associated with overtreatment have supported the introduction of active surveillance as a management option. Multiple guidelines now endorse active surveillance for low-risk PCa, although concerns regarding biopsy under-
sampling and under-staging have limited its acceptance. The development and validation of biopsy-based genomic assays have been published and presented, demonstrating a more accurate risk assessment at the time of diagnosis.
One reason for the current screening challenges in PCa is the lack of a genomic marker that helps to determine the risk of disease progression at the time of initial diagnosis. Most urologists have experience in treating men with apparent low-risk PCa who, upon RP, are found to have higher-grade cancers, which are a surrogate for a higher risk of progression and death.
Prostate Cancer Awareness and Education
The 1980s and 1990s marked the founding of several national education and advocacy groups focused on PCa screening and treatment. These include Patient Advocates for Advanced Cancer Treatment, the Prostate Cancer Education Council, Man to Man, Us TOO! International, and other groups.
A number of these are notable for their efforts to promote PCa screening: The nonprofit Prostate Conditions Education Council (PCEC; www.prostateconditions.org), founded in 1989, is the leading resource for information on prostate health in the United States. A consortium of physicians and scientists, health educators, and prostate cancer advocates, the Council is the founder and coordinator of the national Prostate Cancer Awareness Week Program, and with its Screening Site Partners has screened about 5 million men for PCa across the US.
In the summer of 2000, Arlene Mulder, former Mayor of Arlington Heights, Illinois, founded the Mayors’ Coalition for Prostate Cancer Awareness and Education, to further promote PCa awareness and encourage screening. The same year, the United States Conference of Mayors designated September as Prostate Cancer Awareness Month, the first major national organization to do so. To date, 154 mayors have joined the coalition. In 2002, President George W. Bush officially designated September as National Prostate Cancer Awareness Month.
Cost vs Value
A substantial clinical and economic burden is associated with the diagnosis and treatment of PCa in the US and worldwide. All men diagnosed with PCa and their physicians face the challenge of deciding whether to choose definitive therapy-which has a high cure rate but potentially significant complications-or monitor the disease through active surveillance or watchful waiting. Among the physician, payor, and healthcare system stakeholders in the US and globally there is widespread concern that the rate of treatment for PCa is too high, especially since PCa in many men is often indolent, with little malignant potential. However, there is no question that aggressive therapy is indicated in a large subset of men with aggressive PCa. In our opinion, there is benefit to focusing our treatment on men with high-risk PCa.
Appropriate incorporation of biomarkers into clinical practice can have a positive economic impact. For example, a budget impact study of ConfirmMDx identified men who might avoid unnecessary repeat prostate biopsies, thereby reducing overall healthcare spending.[25] By using the appropriate biomarkers, we can reduce unnecessary biopsies and help men to make more effective clinical decisions about their individual plan of care, which should translate into economic savings.
Conclusion
PCa biomarkers hold tremendous promise for assisting clinicians in improving risk assessment, reducing overtreatment, and providing more selective therapy for patients with high-risk disease. In the last 2 years, a number of new, exciting biomarkers have emerged that offer the opportunity to assist clinicians in determining when to biopsy, whom to re-biopsy, and how to assist patients in their treatment decisions. If third-party payors are to support reimbursement, there will be a need to demonstrate an actionable outcome that occurs and affects patient care.
Financial Disclosure:Dr. Crawford serves on the advisory boards of Dendreon, Ferring, Genomic Health, Janssen, MDxHealth, and Myriad. He is also a speaker for Genomic Health, MDxHealth, and Myriad. Dr. Shore is a consultant to Genomic Health, MDxHealth, and Myriad. Dr. Ventii has no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
References:
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30.
2. Warren JL, Yabroff KR, Meekins A, et al. Evaluation of trends in the cost of initial cancer treatment. J Natl Cancer Inst. 2008;100:888-97.
3. Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28:1117-23.
4. Etzioni R, Penson DF, Legler JM, et al. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94:981-90.
5. National Cancer Institute. SEER cancer statistics. Table 1.18: Lifetime risk (percent) of dying from cancer by site and race/ethnicity. Males, total U.S., 2007-2009. Available from: http://seer.cancer.gov/csr/1975_2009_pops09//results_merged/topic_lifetime_risk.pdf. Accessed January 8, 2014.
6. Howlader N, Noone AM, Krapcho M, et al (eds). SEER Cancer Statistics Review, 1975–2010, National Cancer Institute. Bethesda, MD. Available from: http://seer.cancer.gov/csr/1975_2010. Accessed January 8, 2014.
7. National Cancer Institute. SEER cancer statistics. Table 1.15. Lifetime risk (percent) of being diagnosed with cancer by site and race/ethnicity. Males, total U.S., 2007-2009. Available from: http://seer.cancer.gov/csr/1975_2009_pops09//results_merged/topic_lifetime_risk.pdf. Accessed January 8, 2014.
8. Cooperberg MR, Vickers AJ, Broering JM, Carroll PR. Comparative risk-adjusted mortality outcomes after primary surgery, radiotherapy, or androgen-deprivation therapy for localized prostate cancer. Cancer. 2010;116:5226-34.
9. Lu-Yao GL, Albertsen PC, Moore DF, et al. Outcomes of localized prostate cancer following conservative management. JAMA. 2009;302:1202-9.
10. Carter HB, Albertsen PC, Barry MJ, et al. Early detection of prostate cancer: AUA guideline. J Urol. 2013;190:419-26.
11. Barbieri CE, Bangma CH, Bjartell A, et al. The mutational landscape of prostate cancer. Eur Urol. 2013;64:567-76.
12. Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214-20.
13. Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156-61.
14. Chou R, Croswell JM, Dana T, et al. Screening for prostate cancer: a review of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2011;155:762-71.
15. National Cancer Institute. Prostate-specific antigen (PSA) test. Available from: http://www.cancer.gov/cancertopics/factsheet/detection/PSA. Accessed January 8, 2014.
16. Andriole GL, Crawford ED, Grubb RL 3rd, et al. Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J Natl Cancer Inst. 2012;104:125-32.
17. Crawford ED, Grubb R 3rd, Black A, et al. Comorbidity and mortality results from a randomized prostate cancer screening trial. J Clin Oncol. 2011;29:355-61.
18. Schröder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981-90.
19. Stephan C, Vincendeau S, Houlgatte A, et al. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem. 2013;59:306-14.
20. de la Taille A, Irani J, Graefen M, et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol. 2011;185:2119-25.
21. Crawford ED, Rove KO, Trabulsi EJ, et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol. 2012;188:1726-31.
22. Stewart GD, et al. Clinical utility of a multiplexed epigenetic gene assay to detect cancer in histopathologically negative prostate biopsies: results of the multicenter MATLOC study. American Urological Association annual meeting. Atlanta, GA; 2012. Abstract LBA6.
23. Trock BJ, Brotzman MJ, Mangold LA, et al. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int. 2012;110:56-62.
24. Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol. 2013;189:1110-6.
25. Aubry W. Budget impact model: epigenetic assay can help avoid unnecessary repeated prostate biopsies, reduce spending. Am Health Drug Benefits. 2013;6:15-24.
26. Robinson K, Creed J, Reguly B, et al. Accurate prediction of repeat prostate biopsy outcomes by a mitochondrial DNA deletion assay. Prostate Cancer Prostatic Dis. 2013;16:398.
27. Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72.
28. Leyten GH, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2012;pii: S0302-2838(12)01345-0.
29. Salami SS, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2013;31:566-71.
30. Chaux A, Peskoe SB, Gonzalez-Roibon N, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543-9.
31. Reid AH, Attard G, Ambroisine L, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678-84.
32. Metamark Genetics. About ProMark. Available from: http://www.metamarkgenetics.com. Accessed January 16, 2014.
33. Klein E, Maddala C, Millward C, et al. Development of a needle biopsy-based genomic test to improve discrimination of clinically aggressive from indolent prostate cancer. Poster presentation at the American Society of Clinical Oncology annual meeting. Chicago, IL; 2012. Abstr 456.
34. Cooperberg M, Simko J, Falzarano S, et al. Development and validation of the biopsy-based genomic prostate score (GPS) as a predictor of high grade or extracapsular prostate cancer to improve patient selection for active surveillance. Presented at the American Urological Association annual meeting. San Diego, CA; 2013. Abstr 2131.
35. Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245-55.
36. Ghadjar P, Zwahlen D, Aebersold DM, Zimmermann F. Postoperative radiotherapy after radical prostatectomy: indications and open questions. Prostate Cancer. 2012;2012:963417.
37. Badani K, Thompson DJ, Buerki C, et al. Impact of a genomic classifier of metastatic risk on postoperative treatment recommendations for prostate cancer patients: a report from the DECIDE study group. Oncotarget. 2013;4:600-9.
38. Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047-53.
39. Chen CL, Mahalingam D, Osmulski P, et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate. 2013;73:813-26.
40. Halabi S, Small EJ, Hayes DF, et al. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate-specific antigen in metastatic prostate cancer: a nested study within CALGB 9583. J Clin Oncol.2003;21:490-5.