New Blood Test for Early Ovarian Cancer in Clinical Trials
BETHESDA, Maryland-A new proteomics blood test for ovarian cancer (developed by researchers at the joint Food and Drug Administration/National Institutes of Health Clinical Proteomics Program) detected all 50 ovarian cancers in a proof-of-principal trial and is now being validated in a major study of recurrence in stage III/IV ovarian cancer.
BETHESDA, MarylandA new proteomics blood test for ovarian cancer (developed by researchers at the joint Food and Drug Administration/National Institutes of Health Clinical Proteomics Program) detected all 50 ovarian cancers in a proof-of-principal trial and is now being validated in a major study of recurrence in stage III/IV ovarian cancer.
The new approach also will be used as a monitoring tool in the first study of imatinib mesylate (Gleevec) in ovarian cancer, a phase II trial expected to open soon.
Emanuel F. Petricoin III, PhD, co-director of the Clinical Proteomics Program, told ONI, "This represents an entirely new approach to detecting cancer because it uses the pattern of proteins in the blood as the discriminator, independent of (and indeed without knowing) the identities of the individual proteins.")
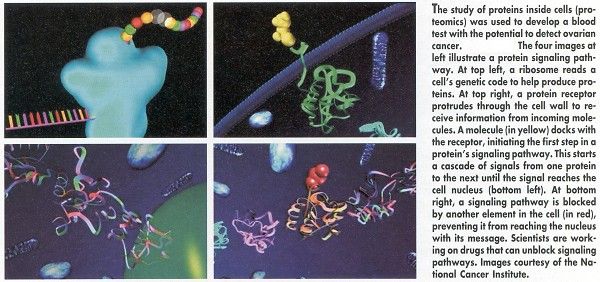
Dr. Petricoin is senior investigator in FDA’s Center for Biologics Evaluation and Research. Co-Director on the NIH side is Lance A. Liotta, MD, PhD, from NCI’s Center for Cancer Research. The study was fast-tracked for publication by The Lancet (359:572-577, 2002) and is available free at the journal’s website www.thelancet.com.
The new test takes a small drop of blood, determines the pattern of proteins present, and compares that with the proteomic patterns known to be associated with cancer. The whole process takes about 30 minutes.
In the pilot study, the algorithm identified a cluster pattern that correctly detected all 50 cancer cases, including 18 stage I cases, in a masked set of samples from a population of women at high risk for ovarian cancer. The pattern also correctly identified 63 of the 66 cases of nonmalignant disease.
The resulting 100% sensitivity and 95% specificity yielded a positive predictive value of 94%far higher than the 20% positive predictive value now possible with CA-125 combined with ultrasound.
The proteomics test represents a dramatic shift from past efforts to find specific biomarkers associated with cancer. "You always have patients for whom single or even multiple biomarkers don’t work," Dr. Petricoin said. "Overall protein patterns are more likely to pick up those cases, and the nature of an artificial intelligence-based, iterative system is that it should get smarter and smarter as time goes on, since it incorporates data from an ever-growing number of cases."
The test was developed, he said, by first training the system on data from patients in whom the outcome was already known. "We used an heuristic adaptation mechanism, which means that the model went back and re-learned on the samples it got wrong in the initial analysis. This makes each subsequent analysis more accurate," he said.
The investigators doubt that the ovarian cancer blood test will ever be a stand-alone diagnostic tool. They expect it to be useful for screening women and deciding who should undergo more extensive examination, and to be an early warning of cancer recurrence.
Dr. Petricoin envisions a system in which blood samples would be analyzed at local clinical laboratories, the raw mass spectra from each patient’s test sent over the internet to a central laboratory housing the analytical software, and results sent electronically back to the patient’s physician.
Cost a Problem
The proteomics blood test will remain a research tool for the present, in part because of cost. The Ciphergen Biosystems (Freemont, California) mass spectrometry equipment required for generating the protein spectra from blood samples costs about $150,000 and must be used with a special chip.
Ciphergen’s mass spectrometry system is particularly amenable to high throughput analysis, meaning that lots of samples can be analyzed at once, but Dr. Petricoin expects more equipment makers to enter the market now that the government team has shown that the test is feasible. "Now that the gauntlet has been thrown down, we expect more companies to work on developing affordable, reliable analyzers that could be used in routine clinical practice," he said.
The chip also needs to be tweaked a bit. Dr. Petricoin said that the Ciphergen chips his group uses are an excellent research tool but are not suitable for routine clinical use. For one thing, they are not made in a factory that has passed the FDA’s stringent good manufacturing practices guidelines. Translating the research chip into a clinical tool is largely an engineering problem, however, and Ciphergen is already at work on it, he said.
The process also requires Proteome Quest bioinformatics software developed by Correlogic Systems, Inc. (Bethesda, Maryland). Ben A Hitt, PhD, from Correlogic is co-author with Drs. Petricoin and Liotta of the Lancet paper. The research team also includes Drs. Ali M. Ardekani, Peter J. Levine, Vincent A. Fusaro, Seth M. Steinberg, Gordon B. Mills, Charles Simone, David A. Fishman, and Elise C. Kohn.
Validating Study
Dr. Elise Kohn is the principal investigator on a trial currently validating the new proteomics test for detecting relapse of ovarian cancer in patients whose stage III or IV epithelial ovarian cancer is in first clinical remission. The new test will also be used in patients with diagnosed pelvic masses of unknown origin, to develop a noninvasive way to distinguish ovarian cancer from ovarian cysts, leiomyosarcomas, and endometrial fibroids.
The research team has already started developing the databases needed to expand proteomic pattern testing to other cancers. Dr. Petricoin said that the group already has "intriguing data" in prostate, breast, lung, colon, and pancreatic cancer.
Dr. Petricoin predicted that proteomic pattern testing will have a "dramatic impact" on drug development by improving the selection of compounds for testing in preclinical studies and by providing early toxicity and efficacy data to help pharmaceutical companies decide which compounds to bring forward to phase I trials.
"Our goal in starting the Clinical Proteomics Program in 1997 was to meld the proteomics and artificial intelligence fields in a way that would yield immediate clinical benefits," Dr. Petricoin said. "This is the kind of problem that a government project with no ethical conflicts of interest is well situated to pursue."
As with the Human Genome Project, proteomic profiles generated by the proteomics program will be posted on the Web and freely available to other researchers.
"I would also like to make a plea here to oncologists, pathologists, and other clinicians to hang onto clinical specimens," Dr. Petricoin said. "Freeze cells and tissues. Store them away when you can. Let us know you have them. The development of reliable proteomic pattern tests will be greatly helped if we can get samples from physicians in private practice in addition to the established banks of research samples."