Circulating Tumor DNA as a Predictive Biomarker for Clinical Outcomes With Margetuximab and Pembrolizumab in Pretreated HER2-Positive Gastric/ Gastroesophageal Adenocarcinoma
Yoon-Koo Kang, MD, PhD
- Hope E. Uronis, MD, MHS
- Keun-Wook Lee, MD, PhD
- Matthew C. H. Ng, MD
- Peter C. Enzinger, MD
- Se Hoon Park, MD, PhD
- Philip J. Gold, MD
- Jill Lacy, MD
- Howard S. Hochster, MD
- Sang Cheul Oh, MD, PhD
- Yeul Hong Kim, MD, PhD
- Kristen A. Marrone, MD
- Ronan J. Kelly, MD
- Rosalyn A. Juergens, MD
- Jong Gwang Kim, MD, PhD
- Thierry Alcindor, MD, MSc
- Sun Jin Sym, MD, PhD
- Eun-Kee Song, MD, PhD
- Cheng Ean Chee, MD
- Yee Chao, MD, PhD
- Sunnie Kim, MD
- Do-Youn Oh, MD, PhD
- Jennifer Yen, PhD
- Justin I. Odegaard, MD, PhD
- Errin Lagow, PhD
- Daner Li, MS
- Jichao Sun, PhD
- Patrick Kaminker, PhD
- Paul A. Moore, PhD
- Minori Koshiji Rosales, MD, PhD
- Haeseong Park, MD, MPH
Daniel V. T. Catenacci, MD, and colleagues present findings from a study of circulating tumor DNA as a predictive biomarker for gastric and gastroesophageal cancer.
ABSTRACT
PURPOSE. To assess the ability of circulating tumor DNA (ctDNA)-based testing to identify patients with HER2 (encoded by ERBB2)-positive gastric/gastroesophageal adenocarcinoma (GEA) who progressed on or after trastuzumab-containing treatments were treated with combination therapy of anti-HER2 and anti–PD-1 agents.
METHODS. ctDNA analysis was performed retrospectively using plasma samples collected at study entry from 86 patients participating in the phase 1/2 CP-MGAH22-05 study (NCT02689284).
RESULTS. Objective response rate (ORR) was significantly higher in evaluable ERBB2 amplification–positive vs – negative patients based on ctDNA analysis at study entry (37% vs 6%, respectively; P = .00094). ORR was 23% across all patients who were evaluable for response. ERBB2 amplification was detected at study entry in 57% of patients (all HER2 positive at diagnosis), and detection was higher (88%) when HER2 status was determined by immunohistochemistry fewer than 6 months before study entry. ctDNA was detected in 98% (84/86) of patients tested at study entry. Codetected ERBB2-activating mutations were not associated with response.
CONCLUSIONS. Current ERBB2 status may be more effective than archival status at predicting clinical benefit from margetuximab plus pembrolizumab therapy. ctDNA testing for ERBB2 status prior to treatment will spare patients from repeat tissue biopsies, which may be reserved for reflex testing when ctDNA is not detected.
KEYWORDS. gastric/gastroesophageal adenocarcinoma; HER2; ctDNA; and margetuximab plus pembrolizumab
Oncology (Williston Park). 2023;37(4):176-183.
DOI: 10.46883/2023.25920992.
BACKGROUND
The human epidermal growth factor receptor 2 (HER2) protein is a receptor tyrosine-protein kinase, encoded by erythroblastic oncogene B2 (ERBB2), normally involved in the proliferation and division of cells. The ERBB2 gene is often amplified and overexpressed in solid tumors, and solid tumors that are HER2-positive are aggressive. In patients with HER2-positive solid tumors, HER2-targeted therapies have become the standard of care in multiple tumor types, including breast, gastric and gastroesophageal (including gastric/gastroesophageal adenocarcinoma [GEA]), salivary, and colorectal cancers. In GEA in particular, HER2-targeted therapies are associated with improved outcomes; however, patients eventually progress, often due to the loss of ERBB2 amplification and subsequent loss of dependence on the ERBB2- signaling pathway.1,2 As subsequent treatment options include both alternative HER2-targeted and nontargeted therapies, it is critical to distinguish between patients whose tumors retain HER2 dependence and those that are HER2 independent; therefore, repeated testing is necessary to determine ERBB2 amplification status. Traditional assessment of ERBB2 amplification—the most common cause of HER2 dependence in GEA—requires tissue samples, but repeated biopsies are not feasible for many patients. Moreover, HER2 status has been reported to vary among different regions in a tumor in an individual patient with GEA,3-5 so HER2 status cannot necessarily be captured by a single tissue biopsy.
Cell-free DNA (cfDNA)-based liquid biopsies have the potential to address these limitations by assessing genomic information from across the entire tumor volume from a peripheral blood draw. The feasibility of such assessment specifically for tumor-derived ERBB2 amplification has been previously demonstrated in multiple indications, including in GEA, both in terms of concordance with tissue-based testing and of predicting the clinical benefit of HER2-targeted therapies.3-13 Moreover, circulating tumor–derived DNA (ctDNA) levels have also been shown to correlate with disease burden in GEA,4 and changes in ctDNA levels are able to predict clinical benefit from both HER2-targeted therapies7,10,12,14-16 and immunotherapies.17-20 cfDNA is present in plasma and serum, and it is seen in higher quantities in patients with cancer.21 ctDNA is the fraction of cfDNA shed into circulation by apoptotic and necrotic tumor cells in patients with cancer.22 Approaches for detection of the ctDNA fraction in cfDNA include targeting of defined changes in single alterations or use of next-generation sequencing–based comprehensive genomic profiling (including targeted and whole exome/genome sequencing) to interrogate all possible aberrations in DNA; detection of the ctDNA fraction also permits assessment of clonal differences in tumor cell populations.23
Margetuximab is approved in combination with chemotherapy for patients with HER2-positive metastatic breast cancer who have received 2 or more prior anti-HER2 regimens, at least 1 of which was for metastatic disease. Margetuximab is also being investigated in patients with HER2-positive GEA. Margetuximab is an anti-HER2 monoclonal antibody that is designed to have increased binding to the activating fragment crystallizable (Fc) receptor FcγRIIIA (CD16A) and decreased binding to the inhibitory Fc receptor FcγRIIB (CD32B) compared with trastuzumab, delivering more potent antitumor responses via antibody-dependent cellular cytotoxicity.6,24 Pembrolizumab is an anti–PD-1 monoclonal antibody approved in combination with trastuzumab-, fluoropyrimidine-, and platinum-containing chemotherapy for the first-line treatment of patients with locally advanced, unresectable or metastatic HER2-positive GEA.7
In this study, we assessed the ability of cfDNA-based testing to identify patients with HER2-positive GEA who progressed on or after trastuzumab-containing treatments and who were likely to benefit from combination therapy of anti-HER2 and anti–PD-1 agents, based on pretreatment presence of ERBB2 amplification and additional ctDNA-based biomarkers.
METHODS
Patients in CP-MGAH22-05 ctDNA study
We performed a retrospective ctDNA analysis on prospectively collected samples from patients with previously treated, locally advanced, unresectable or metastatic HER2-positive GEA in a single-arm, open-label, phase 1b/2, dose-escalation and cohort expansion study (CP-MGAH22-05; NCT02689284).6 Patients must have had histologically proven unresectable locally advanced or metastatic HER2-positive GEA and received prior treatment with trastuzumab. Tissue biopsy HER2 status was based on local testing as described previously.6 HER2 positivity was assessed using immunohistochemistry (IHC), defined as IHC3 positive or IHC2 positive, and amplified fluorescence in situ hybridization (FISH), defined as a HER2 to chromosome enumeration probe 17 ratio of 2.0 or more.25 Tissue biopsy PD-L1 status, although not used for enrollment, was determined centrally by IHC, and PD-L1 positivity was defined as a combined positive score of 1 or greater.6 In the dose-escalation phase, 9 patients were treated: 3 received margetuximab at 10 mg/kg intravenously (IV) plus pembrolizumab at 200 mg IV every 3 weeks, and 6 received the recommended phase 2 dose (RP2D) of margetuximab at 15 mg/kg plus pembrolizumab at 200 mg IV every 3 weeks. An additional 86 patients were enrolled in the phase 2 cohort expansion and received the RP2D. Plasma ctDNA was available from 86 of the 95 patients enrolled (the phase 2 cohort expansion), of whom 83 received the RP2D of margetuximab and pembrolizumab and 3 received margetuximab at 10 mg/kg and pembrolizumab.
The study was conducted according to International Conference on Harmonization Guideline for Good Clinical Practice and all applicable local and national regulations and ethical principles in accordance with the Declaration of Helsinki. All patients provided written informed consent. The protocol and the informed consent document were reviewed and approved by the institutional review board or independent ethics committee of each participating center before study initiation.
ctDNA analysis
Plasma samples collected from patients in the phase 2 cohort expansion were tested for ctDNA using the Guardant360 CDx.26 Guardant360 is an FDA-approved27 test that detects single nucleotide variants (SNVs; 73 or 74 genes), copy number amplifications (19 genes), insertion-deletion alterations (23 genes), and fusions (6 genes) in plasma from patients with solid tumors (full list of genes provided in the Supplement). ctDNA sequencing data was analyzed on the Guardant360 bioinformatics pipeline as previously described.4,12 Briefly, gene-level copy number alterations were determined after probe-level unique molecule normalization, diploid baselining, background noise correction, and comparison with established reporting decision thresholds.12 ctDNA detection was defined as presence of 1 or more somatic alterations per patient sample.
Statistical analysis
The objective response rate (ORR) was defined as the proportion of patients who achieved a confirmed complete response (CR) or confirmed partial response (PR). The clinical benefit rate (CBR) was defined as the proportion of patients who achieved a confirmed CR or confirmed PR or stable disease. The CBR was not a prespecified trial end point and was later selected as a marker of clinical benefit because this provided a larger sample size than the population of responders (CR/PR). Categorical data were summarized by the number and percentage of patients for each variable. Fisher’s exact test was performed to compare binary end points between 2 groups divided according to ORR or CBR. A P value less than .05 was considered significant.
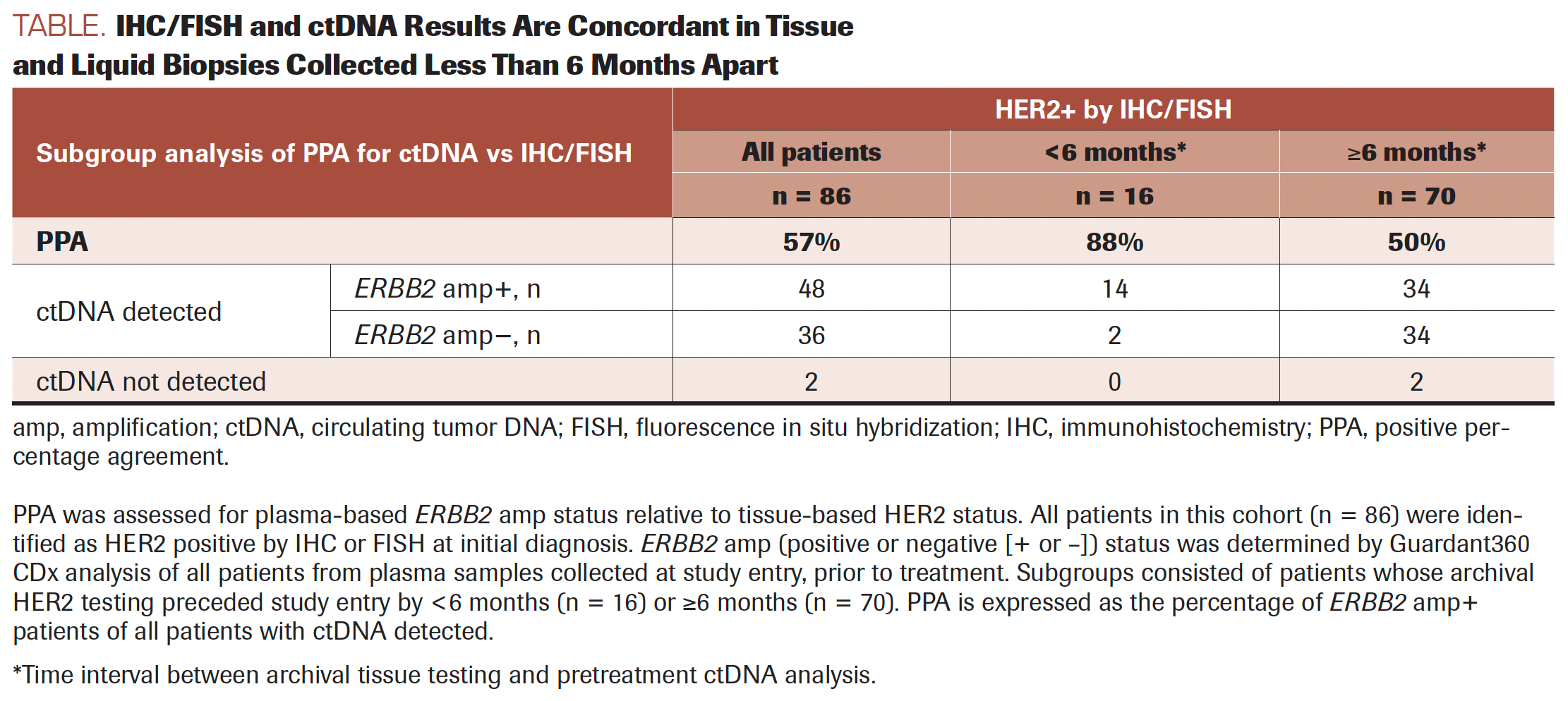
TABLE. IHC/FISH and ctDNA Results Are Concordant in Tissue and Liquid Biopsies Collected Less Than 6 Months Apart
RESULTS
Concordance between ERBB2 amplification detection using ctDNA and IHC/FISH
A total of 86 patients, 83 of whom received the RP2D of margetuximab (15 mg/kg) plus pembrolizumab and 3 of whom received margetuximab (10 mg/ kg) plus pembrolizumab, had ctDNA testing at study entry, after progression on standard-of-care HER2-targeted therapy. At baseline, 84 of 86 patients (98%) had more than 1 reportable genomic alteration detected using ctDNA. As reported in the first publication from this trial,6 ERBB2 gene amplification was detected at study entry in ctDNA in 57% (48 of 84) of patients with ERBB2 (HER2) amplification detected at the time of diagnosis using IHC or FISH (Table). The apparent lower rate of ERBB2 gene amplification detection using ctDNA may reflect the loss of ERBB2 amplification after progression on trastuzumab, which has been reported in 30% to 60% of initially HER2-positive gastric cancer cases, or it may reflect false-negative ctDNA results due to low tumor shedding.4,28 To address the first possibility, we first asked whether the ERBB2 amplification detection rate was higher among patients with HER2 IHC status determined from repeat biopsies collected post progression on trastuzumab. HER2 positivity for 9 of 86 patients had been determined from repeat tissue biopsies; these were not required but were performed following trastuzumab failure. This subset of 9 patients permitted a small but direct comparison of HER2 IHC and ERBB2 amplification status due to lack of intervening trastuzumab treatment and short time interval between tissue and liquid biopsy (median, 19 days; range, 1-146). Amplified ERBB2 was detected in ctDNA at study entry from 8 of the 9 patients for whom repeat tissue biopsies were HER2 positive. Maximum variant allele frequencies (maxVAFs) were 3% or higher for these 8 ERBB2 amplification-positive cases vs 0.16% for the single ERBB2 amplification-negative case, suggesting that the negative case was likely a false negative due to low tumor shedding.
Given the small sample size of paired tissue and liquid biopsies without intervening trastuzumab, we also asked whether the ERBB2 detection rate was higher in patients with a short time interval between tissue testing and plasma collection for ctDNA analysis. True loss of ERBB2 amplification is a time-dependent biological process, and this should result in a greater difference over time, whereas false negatives by ctDNA should be time-independent. The time between ctDNA testing at study entry and IHC/FISH at diagnosis ranged from a few days to more than 6 years, with some patients having progressed on trastuzumab with multiple combinations of chemotherapy between diagnosis and study entry. Across this range, tissue-ctDNA concordance demonstrated a strong dependence on the length of the interval between tests; the positive percentage agreement for tests performed less than 6 months apart was 88% (14/16) vs 50% (34/68) for tests performed 6 months apart or more (P = .0099).
Six months provided a sufficient sample size for the comparison and is aligned with the median progression-free survival (PFS) reported from the phase 3 ToGA trial (NCT01041404) of trastuzumab plus chemotherapy in patients with treatment-naive advanced GEA.29 To address the question of whether ERBB2 detection rate was higher among patients with higher tumor shedding, we explored maxVAFs across all 84 patients with ctDNA detected. The dependency of gene amplification detection on high tumor fraction is widely known, and the Guardant360 method for copy number assessment has been described previously, but a distinct ctDNA maxVAF threshold does not exist.27 For simplicity and the purpose of our analysis, ctDNA maxVAFs below 1% were used to define low tumor shedding. Among patients with ctDNA detected, 42% (15 of 36) of ERBB2 amplification–negative cases were associated with low tumor shedding (<1% ctDNA maxVAF) vs 10% (5 of 48) of ERBB2 amplification–positive cases (Fisher exact test, P = .0015), confirming the relationship between tumor fraction and copy number. However, all but 1 patient with ERBB2 amplification–negative tumors also had intervening trastuzumab-based treatment and a long time interval (median, 451 days; range, 167-2429) between HER2 IHC and ctDNA analysis; this makes it difficult to distinguish false negatives (due to low tumor shedding) from true negatives (due to HER2 loss).
FIGURE 1.
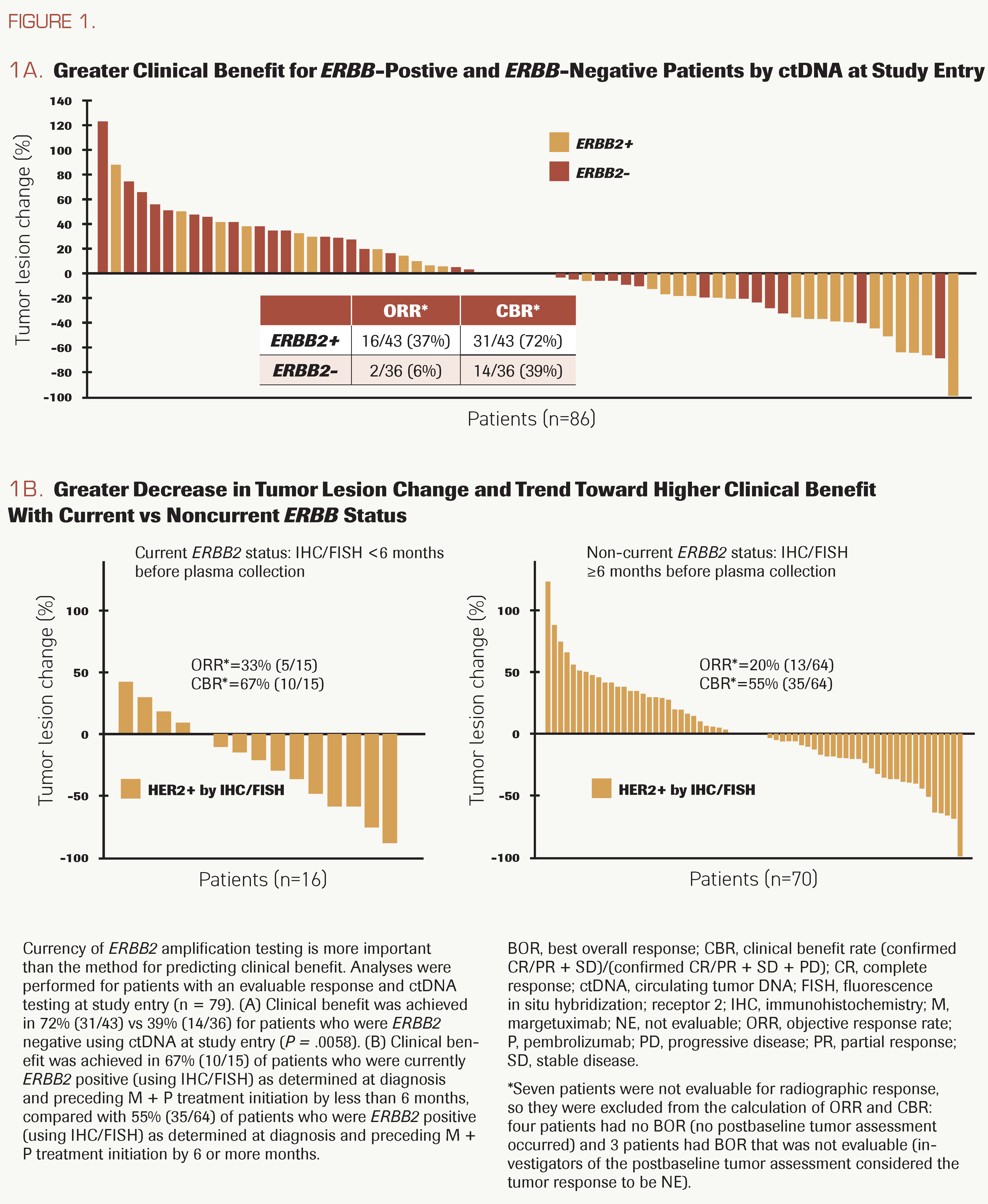
Current ERBB2 status predicts response
Rates of objective response and clinical benefit from margetuximab and pembrolizumab combination therapy in patients from the CP-MGAH22-05 study were reported previously.6 Updated treatment duration and outcome data became available in July 2021. Among the 79 patients who were evaluable for response and had ctDNA testing at study entry, ORR and CBR were significantly higher in patients who were ERBB2 amplification positive than in those who were negative: ORR, 37% (16 of 43) vs 6% (2 of 36); P = .00094; CBR, 72% (31 of 43) vs 39% (14 of 36); P = .0058 (Figure 1A). To assess the importance of timing independent of the test method, we also compared the ORR between cases that were identified as HER2-positive less than 6 months before study entry vs the entire cohort, in which all cases were identified as HER2-positive at diagnosis or after progression on trastuzumab and in some cases years before study entry. Although ORR was numerically higher in cases with current HER2-positive status, the difference was not significant, as the full cohort likely includes cases with HER2 loss due to prior trastuzumab-based treatment (Figure 1B). These results suggest that the timing of HER2 or ERBB2 amplification testing is more important than the method (IHC or ctDNA) for predicting clinical benefit.
VAFs and maxVAF
Baseline maxVAF values were explored for potential association with clinical outcomes. Negative correlations between maxVAF and PFS and between maxVAF and overall survival (OS) exist (r = –0.229 and r = –0.270, respectively) and are significant (P = .0394 and P = .014, respectively), indicating that lower tumor shedding, based on maxVAF as a proxy, is associated with longer PFS and OS. Numerically lower maxVAF at baseline (defined as ≤10% for the purpose of the analysis) was associated with longer median PFS and OS than higher maxVAF at baseline (defined as >10%), although the associations with PFS (median of 84 days vs 43 days, respectively) or OS (median of 423 and 221 days, respectively) were not statistically significant. A similar analysis was performed to ask whether longer PFS or OS were associated with baseline maxVAF of 1% or less. No significant differences in PFS or OS were observed between patients with baseline maxVAF greater than 1% vs 1% or less. Median PFS for patients with baseline maxVAF of 1% or less was 99 days vs 81.5 days for maxVAF greater than 1% (log rank test, P = .87). Median OS for maxVAF greater than 1% was 345 days, vs 404 days for maxVAF of 1% or less (log rank test, P = .09).
FIGURE 2. Biomarker and Response
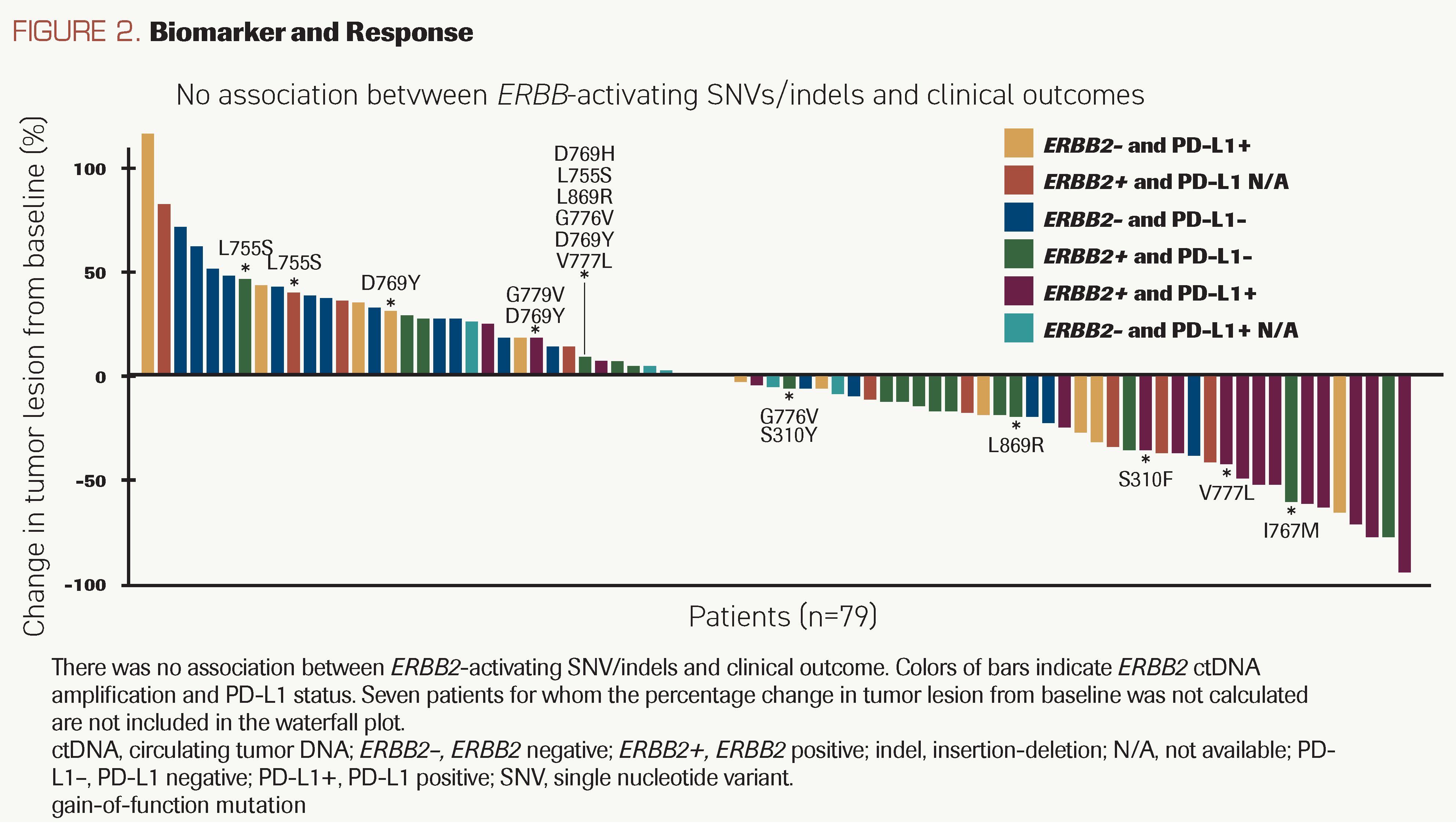
Co-occurring ERBB2 SNVs and clinical response
Using ctDNA analysis, all patients were assessed for mutations in 74 genes commonly mutated in cancer, in addition to ERBB2 amplification. Gain-of-function ERBB2 SNVs D769Y/H, S310F/Y, L755S, and V777L, may confer resistance to traditional antibody-based anti-HER2 therapies and were detected in 19% (16/84) of patients, all but 1 of whom retained ERBB2 amplification (Figure 2; Supplement Figure 2). In the 13 patients evaluable for response, CBR was 69% (9 of 13), suggesting that the drug combination is effective in patients with both ERBB2 amplification and ERBB2-activating SNVs.
FIGURE 3.
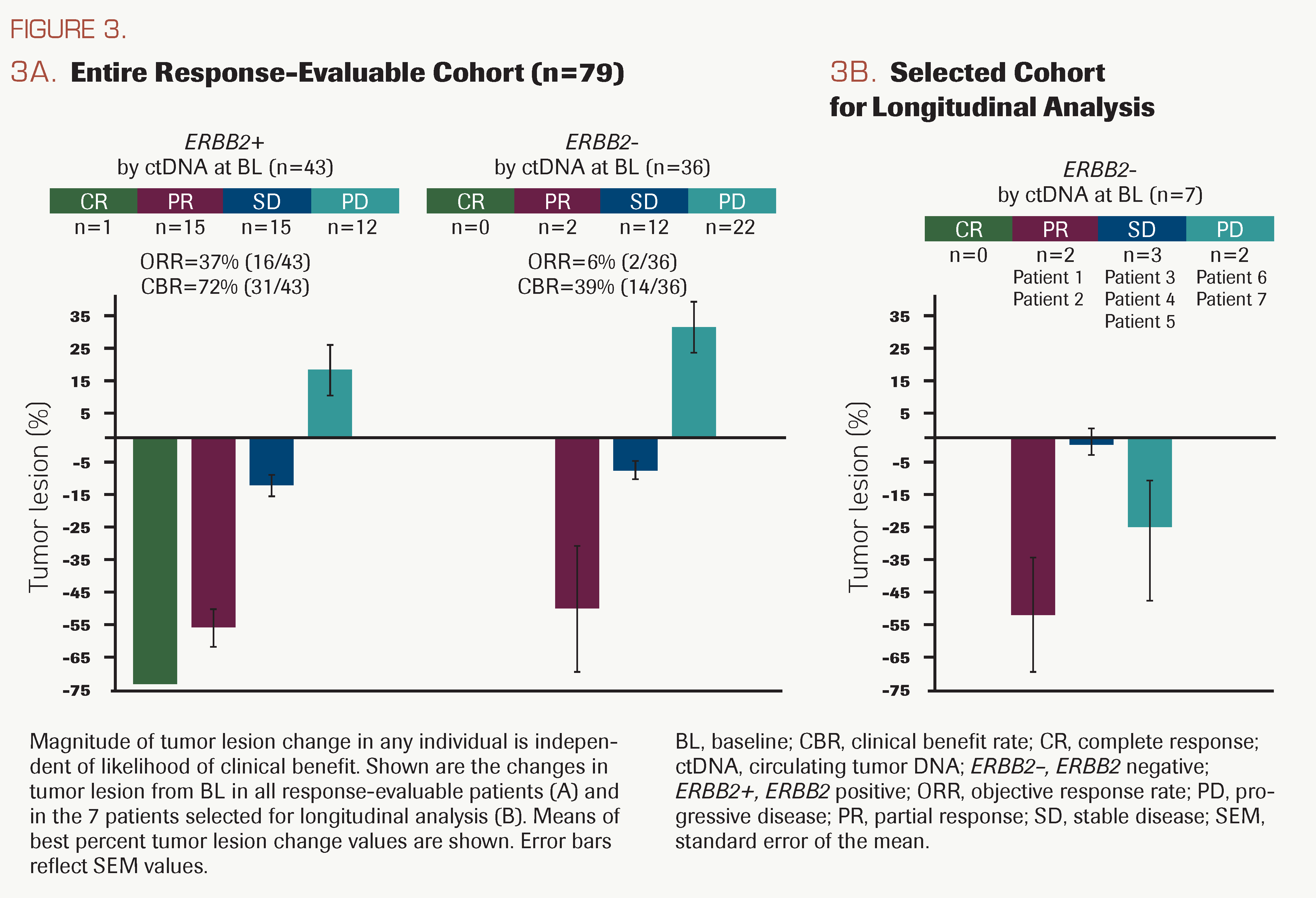
Association between baseline ctDNA ERBB2 amplification status and response
The ORR was significantly higher in patients who were ERBB2 amplification positive vs negative at study entry (37% vs 6%, P = .00094; Figure 1A); however, the magnitude of the change in tumor lesion size was similar between patients who were ERBB2 amplification positive and those who were ERBB2 amplification negative at study entry (Figure 3A). Tumor shrinkage also was observed in a subset of 7 patients who were ERBB2 amplification negative at study entry and experienced long treatment duration (Figure 3B). Of the 36 patients identified as being ERBB2 amplification–negative at study entry, 7 were selected for analysis of ctDNA at later treatment time points.
Longitudinal analysis of ctDNA from selected patients
In the 7 selected patients for on-treatment ctDNA analysis, the best overall response varied, but they collectively had long treatment durations; PFS and OS were comparable with those achieved for patients who were ERBB2 amplification positive at study entry. Statistical analysis of longitudinal ctDNA data vs response or outcomes was not possible due to the small sample size, but details from each case are summarized to speculate on potential explanations for the longer treatment durations of this subgroup. Longitudinal ctDNA fractions of these 7 patients are plotted graphically in Supplement Figure 1, and additional details are provided in the Supplement Table and the Supplement. In 4 of these patients (2, 3, 4, and 6), there was a net decrease or minimal net change in ctDNA from baseline to cycle 4 day 1 (C4D1), consistent with stabilization or tumor lesion shrinkage (best percentage tumor lesion change) and compatible with initial treatment effect. Among the 3 patients with a net increase in ctDNA from baseline to C4D1, the best percentage tumor lesion change was a small decrease (≤10%) for 2 patients (5 and 7) and a somewhat larger decrease (33%) for patient 1. For patients 5 and 7, a potential explanation for the net increase in ctDNA is the presence of additional tumors at C4D1 that were not yet measurable by RECIST v1.1 but, in the aggregate, meaningfully contributed to ctDNA levels, as has been previously reported.14-18,30-32 For patient 1, who achieved PR, the apparent increase in ctDNA level was unexpected and should be viewed with caution, as the C4D1 sample input was below the range that is supported for clinical testing. Despite long treatment duration, subclonal KRAS and TP53 SNVs at baseline emerged as dominant clones by end of treatment (EOT) and by C4D1, respectively, for 2 patients (6 and 7) whose best confirmed response was progressive disease (PD) per RECIST 1.1. Patient 6 developed a new lesion with substantial main tumor shrinkage (Figure 3B). The tumor of patient 7 enlarged at first assessment but shrunk during subsequent treatment that was received after radiographic PD (Supplement Figure 1; Supplement Table). It is noteworthy that patient 7 was ERBB2 gene amplification negative before treatment but became positive after radiographic PD (C4D1) and at EOT.
DISCUSSION
Trastuzumab in combination with chemotherapy is the standard therapy for treatment naive HER2-positive GEA; however, most patients who initially benefit from trastuzumab-based therapy develop resistance. As the potential therapeutic options for such patients include both additional HER2-targeted and nontargeted therapies, it is critical to assess whether patients’ tumors retain or have lost their HER2 dependence.
In this study, we assessed the ability of ctDNA testing for ERBB2 amplification to identify previously treated patients with HER2-positive GEA who are likely to benefit from the combination of the anti-HER2 agent margetuximab and the anti–PD-1 agent pembrolizumab. Of all evaluable patients, all of whom were HER2 positive based on tissue biopsies at diagnosis or after progression on trastuzumab-based therapy, only 57% of patients retained detectable ERBB2 amplification at study entry, consistent with previous reports of loss of HER2 positivity, which ranged approximately from 30% to 60% among previously treated patients with HER2-positive GEA.33-36 Although a limitation of this study was that tissue-based biomarker information was established using diagnostic specimens obtained before and after trastuzumab treatment, ERBB2 gene amplification in ctDNA was nevertheless associated with HER2 protein expression as measured using IHC in a time-dependent manner, with more recent tissue testing demonstrating higher concordance regardless of testing modality. Importantly, we observed that patients with retained ERBB2 amplification as measured using ctDNA at the time of study entry were more likely to experience an objective response from combined anti-HER2/anti–PD-1 therapy, consistent with previous reports of other anti-HER2 agents.4,11,13 However, for patients with ERBB2-negative disease assessed by ctDNA, potentially because of low ctDNA shed, a biopsy would be preferable to further evaluate HER2 status.
On-treatment ctDNA dynamics have been reported as predictive of benefit from both anti-HER2 therapies7,10,12,14-16 and immune checkpoint blockade.17-20 We analyzed on-treatment ctDNA in selected patients without ctDNA ERBB2 amplification at baseline, and although this analysis is limited by small sample size and lack of correlation to the primary end point of the clinical trial, we found that ctDNA VAF dynamics generally suggested a nonprespecified exploratory end point of clinical response in those patients. Validation in a larger, adequately powered cohort is needed to assess the benefit. In some cases, on-treatment changes in maxVAFs were more powerful predictors of both response and progression than radiographic response alone, consistent with previous reports of pembrolizumab treatment in HER2-positive gastric cancer.14,17
Known hotspot mutations reported in all cancers were observed in amplified ERBB2 alleles in nearly 20% (16 of 84) of patients in this study of previously treated patients with HER2-positive GEA, including S310F, V777L, L755S, and D769Y, which are predicted to be driver mutations.37,38 ERBB2 S310F/Y mutations in particular have been associated with poor response to trastuzumab in lung cancer; however, we observed no diminution of efficacy in such patients with ERBB2 comutated GEA, indicating that the combination of margetuximab plus pembrolizumab remains effective in patients with GEA with both ERBB2 amplification and ERBB2-activating SNVs.39
An important limitation of this study is that concurrent ctDNA and tissue testing results were available only for a small subgroup of patients (tissue testing was performed at diagnosis for most of the cohort). As such, these data cannot inform a sufficiently powered comparison of testing methods; however, our primary results do suggest that the currency of testing is more important than the testing modality. In clinical practice, it is likely that the most appropriate testing modality may vary owing to patient- and situation-specific factors, such as access to current tissue and turnaround time.
The longitudinal ctDNA analysis described here for a small subset of patients who were ERBB2 amplification negative at study entry revealed the emergence of additional alterations on treatment, which could be assessed for potential associations with response in a larger, adequately powered study. Based on The Cancer Genome Atlas classification, gastric cancer–overexpressed HER2 is more frequent in chromosomal instability subtypes, which show marked aneuploidy and focal activation of the receptor tyrosine kinases–RAS (RTK-RAS) pathway and high frequency of TP53 mutations.40 A previous study also showed that 5 genes (CCNE1, PIK3CA, KRAS, CDK4, and CDK6) were concomitantly co-amplified and that some genes, such as TP53, CDKN2A, KRAS, KIT, and PIK3CA, were concomitantly comutated in HER2-positive gastric cancer.41 Acquired mutations in KRAS (G12D and T35A), NF1 (N1503S), and PIK3CA (E542K and S1008T) and co-amplifications of BRAF, KRAS, PIK3CA, and FGFR1 are believed to likely represent mechanisms of resistance to anti-HER2 therapy.4
In conclusion, our results suggest that a personalized treatment strategy based on testing for current ERBB2 amplification status should be further explored for optimal selection of therapy in patients with GEA who are progressing on trastuzumab-based therapy, to successfully improve outcomes with targeted therapeutics in this disease. As a first choice of modality to identify persistent HER2- driven disease, ctDNA analysis will save patients from receiving repeat biopsies and will expand testing access to patients for whom repeat biopsies are infeasible. However, for patients with no ctDNA detected or patients with an ERBB2 amplification–negative result and low tumor shedding, reflex testing to repeat tissue biopsy may be useful to further evaluate HER2 status. As such, the difficulty, invasiveness, and inconvenience of obtaining post-progression tissue biopsies, coupled with the predictive value of ERBB2 amplification as assessed using ctDNA, support the practice of liquid biopsy molecular profiling to detect retention of HER2-driven disease for the continuation of HER2-directed therapies in patients with HER2-positive GEA who were previously treated with anti-HER2 agents. The findings from this report corroborate the prudence of this strategy.
AUTHOR AFFILIATIONS:
Daniel V. T. Catenacci, MD1*; Yoon-Koo Kang, MD, PhD2*; Hope E. Uronis, MD, MHS3; Keun-Wook Lee, MD, PhD4; Matthew C. H. Ng, MD5; Peter C. Enzinger, MD6; Se Hoon Park, MD, PhD7; Philip J. Gold, MD8; Jill Lacy, MD9; Howard S. Hochster, MD9†; Sang Cheul Oh, MD, PhD10; Yeul Hong Kim, MD, PhD11; Kristen A. Marrone, MD12; Ronan J. Kelly, MD13; Rosalyn A. Juergens, MD14; Jong Gwang Kim, MD, PhD15; Thierry Alcindor, MD, MSc16; Sun Jin Sym, MD, PhD17; Eun-Kee Song, MD, PhD18; Cheng Ean Chee, MD19; Yee Chao, MD, PhD20; Sunnie Kim, MD21‡; Do-Youn Oh, MD, PhD22; Jennifer Yen, PhD23; Justin I. Odegaard, MD, PhD23; Errin Lagow, PhD23; Daner Li, MS24; Jichao Sun, PhD24; Patrick Kaminker, PhD24; Paul A. Moore, PhD24; Minori Koshiji Rosales, MD, PhD24§; and Haeseong Park, MD, MPH25
1The University of Chicago Medical Center, Chicago, IL
2Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea
3Duke University Medical Center, Durham, NC
4Seoul National University College of Medicine, Seoul National University Bundang Hospital, Seongnam, South Korea
5National Cancer Centre Singapore, DukeNUS (Duke University – National University of Singapore) Medical School, Singapore
6Dana-Farber Brigham Cancer Center, Boston, MA
7Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea
8Swedish Cancer Institute, Seattle, WA
9Yale School of Medicine, New Haven, CT
10Korea University Guro Hospital, Seoul, South Korea
11Korea University Anam Hospital, Seoul, South Korea
12Johns Hopkins University, Baltimore, MD
13Baylor University Medical Center, Dallas, TX
14McMaster University, Juravinski Cancer Centre, Hamilton, Ontario, Canada
15Kyungpook National University Chilgok Hospital, Daegu, South Korea
16McGill University Health Centre, Montreal, Quebec, Canada
17Gachon University Gil Medical Center, Incheon, South Korea
18Jeonbuk National University Medical School, Jeonju, South Korea
19National University Cancer Institute, Singapore
20Taipei Veterans General Hospital, Taipei, Taiwan
21Georgetown University Lombardi Comprehensive Cancer Center, Washington, DC
22Seoul National University Hospital, Cancer Research Institute, Seoul National University College of Medicine, Integrated Major in Innovative Medical Science, Seoul National University Graduate School, Seoul, South Korea
23Guardant Health, Redwood City, CA
24MacroGenics, Rockville, MD
25Washington University School of Medicine, St Louis, MO
*Contributed equally.
†Current affiliation: Rutgers Cancer Institute of New Jersey, New Brunswick, NJ.
‡Current affiliation: University of Colorado Cancer Center, Aurora, CO.
§Current affiliation: Sesen Bio, Cambridge, MA.
SUPPORT:
Funding for this study was provided by MacroGenics, Inc.
CORRESPONDING AUTHOR:
Yoon-Koo Kang, MD, PhD
Asan Medical Center
University of Ulsan College of Medicine
88 Olympic-ro 43-gil
Sonpa-gu, Seoul 05505
Republic of Korea
Tel: 82-2-3010-3210
Fax: 82-2-3010-8772
Email: ykkang@amc.seoul.kr
PRIOR PRESENTATION:
There has been no prior presentation of this original research.
DATA SHARING:
All data relevant to the study are included in the article or uploaded in the Supplement.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST:
DVTC reports receiving fees and honoraria from Amgen, Archer, Arcus Biosciences, Astellas Pharma, Basilea Pharmaceutica, Bristol Myers Squibb, Blueprint Medicines, Daiichi Sankyo, Five Prime Therapeutics, Foundation Medicine, Genentech/Roche, Guardant Health, Gritstone Oncology, Lilly, Merck, Natera, Ono Pharmaceutical, Pieris Pharmaceuticals, Pierian Biosciences, QED Therapeutics, Seattle Genetics, Servier Pharmaceuticals, Silverback Therapeutics, Taiho Pharmaceutical, Tempus Labs, and Zymeworks; YKK reports receiving consulting fees from ALX Oncology, Amgen, Bristol Myers Squibb, Dae Hwa Pharmaceutical, MacroGenics, Merck, Novartis, and Zymeworks; HEU reports receiving research funding to their institution from Bristol Myers Squibb, Leap Therapeutics, MacroGenics, and Merck, consulting fees from Bristol Myers Squibb, travel support for an investigator meeting from Bristol Myers Squibb, and personal fees for participation on a data safety monitoring board/advisory board from AstraZeneca; KWL reports receiving consulting fees from Bayer, Bristol Myers Squibb, Daiichi Sankyo, and ISU ABXIS, and research grants to their institution for conducting clinical trials from ABL Bio, ALX Oncology, Astellas Pharma, AstraZeneca, BeiGene, Daiichi Sankyo, Five Prime Therapeutics, Genexine, Green Cross Corp, LSK BioPharma, MacroGenics, Merck KGaA, Merck Sharp & Dohme, MedPacto, OncXerna Therapeutics, Ono Pharmaceutical, Pharmacyclics, Pfizer, Taiho Pharmaceutical, Y-Biologics, and Zymeworks; PCE reports receiving consulting fees from ALX Oncology, Arcus Biosciences, Astellas Pharma, AstraZeneca, Blueprint Medicines, Bristol Myers Squibb, Celgene, Coherus BioSciences, Daiichi Sankyo, Five Prime Therapeutics, IDEAYA Biosciences, Istari Oncology, Legend Biotech, Lilly, Loxo Oncology, Merck, Ono Pharmaceutical, Servier Pharmaceuticals, Taiho Pharmaceutical, Takeda, Turning Point Therapeutics, Xencor, and Zymeworks; JL reports receiving consulting fees for advisory board from Celgene, Deciphera, Guidepoint, Ipsen Pharma, KeyQuest Health, and Merck, and honoraria from the American Society of Clinical Oncology and St. Francis Hospital, HartforD, CT; HSH reports receiving consulting fees from Bristol Myers Squibb, Genentech/ Roche, and Merck; KAM reports receiving consulting fees from AstraZeneca, honoraria for presentations from AstraZeneca and Puma Biotechnology, and fees for participation on a data safety monitoring board/advisory board from Amgen, AstraZeneca, and Mirati Therapeutics; RJK reports receiving consulting fees from Bristol Myers Squibb, payment for expert testimony from Merck, and fees for participation on a data safety monitoring board/advisory board from Astellas Pharma, AstraZeneca, Bristol Myers Squibb, Daiichi Sankyo, Merck, and Novartis; RAJ reports receiving research funding to their institution from AstraZeneca/MedImmune, Bristol Myers Squibb, Debiopharm Group, Merck Sharpe Dohme, Novartis, and Turnstone Bio, honoraria from Amgen, AstraZeneca, Bristol Myers Squibb, Merck Sharp & Dohme, Novartis Canada Pharmaceuticals, Roche Canada, and consulting fees from AbbVie, Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, EMD Serono, Fusion Pharmaceuticals, Jazz Pharmaceuticals, Lilly, Merck Sharpe Dohme, Novartis, Pfizer, Roche Canada, Sanofi/ Regeneron, and Takeda; TA reports receiving research funding to their institution from Astellas Pharma, EMD Serono, and MacroGenics, consulting fees from Bristol Myers Squibb, Merck, and Roche, and fees for participation on a data safety monitoring board/advisory board from Amgen and AstraZeneca; SK reports receiving honoraria from Astellas Pharma and Merck; DYO reports receiving grants from Array BioPharma, AstraZeneca, BeiGene, HANDOK, Lilly, Merck Sharp & Dohme, Novartis, and Servier Pharmaceuticals, and fees for participation on a data safety monitoring board/advisory board from ASLAN Pharmaceuticals, AstraZeneca, Basilea Pharmaceutica, Bayer, BeiGene, Celgene, Genentech/Roche, Halozyme, Merck Serono, Novartis, Taiho Pharmaceutical, Turning Point Therapeutics, and Zymeworks; JY, JO and EL report employment by Guardant Health and receiving stock interests from Guardant Health; DL, JS, PK, and PAM report employment by MacroGenics and receiving stock interests from MacroGenics; MR reports past employment by MacroGenics and employment by Sesen Bio; and HP reports grants to their institution from Adlai Nortye USA, Alpine Immune Sciences, Ambrx, Amgen, Aprea Therapeutics AB, Array BioPharma, Bayer, BeiGene, BJ Bioscience, Bristol Myers Squibb, Daiichi Sankyo, Elicio Therapeutics, EMD Serono, Exelixis, Genentech, Gilead Sciences, GlaxoSmithKline, Gossamer Bio, Hoffman-La Roche, Hutchison MediPharma, ImmuneOncia Therapeutics, Incyte, Jounce Therapeutics, Lilly, MabSpace Biosciences, MacroGenics, MedImmune, Medivation, Merck, Millennium Pharmaceuticals, Mirati Therapeutics, Novartis, OncXerna Therapeutics, Pfizer, PsiOxus Therapeutics, Puma Biotechnology, Regeneron Pharmaceuticals, Repare Therapeutics, Seattle Genetics, Synermore Biologics, Taiho Pharmaceutical, TopAlliance Biosciences, Turning Point Therapeutics, Vedanta Biosciences, and Xencor. No other potential conflicts of interest were reported.
AUTHOR CONTRIBUTIONS:
Conception and design: Daniel V. T. Catenacci, Eun-Kee Song, Minori K. Rosales, Thierry Alcindor, Yoon-Koo Kang, Paul A. Moore
Development of methodology: Daniel V. T. Catenacci, Eun-Kee Song, Minori K. Rosales, Thierry Alcindor
Collection and assembly of data: Daniel V. T. Catenacci, Do-Youn Oh, Eun-Kee Song, Peter C. Enzinger, Jill Lacy, Rosalyn A. Juergens, Keun-Wook Lee, Philip J. Gold, Minori K. Rosales, Sang Cheul Oh, Se Hoon Park, Sun Jin Sym, Sunnie Kim, Thierry Alcindor, Yee Chao, Yeul Hong Kim, Yoon-Koo Kang, Daner Li, Hope E. Uronis, Jennifer Yen, Justin I. Odegaard, Matthew C. H. Ng, Kristen A. Marrone, Howard S. Hochster
Data analysis and interpretation: Daniel V. T. Catenacci, Do-Youn Oh, Eun-Kee Song, Rosalyn A. Juergens, Patrick Kaminker, Philip J. Gold, Minori K. Rosales, Se Hoon Park, Sun Jin Sym, Thierry Alcindor, Yee Chao, Yoon- Koo Kang, Daner Li, Jennifer Yen, Justin I. Odegaard, Errin Lagow, Howard S. Hochster, Paul A. Moore
Writing, review, and/or revision of the manuscript: Daniel V. T. Catenacci, Do-Youn Oh, Eun-Kee Song, Peter C. Enzinger, Jill Lacy, Rosalyn A. Juergens, Ronan J. Kelly, Keun- Wook Lee, Patrick Kaminker, Philip J. Gold, Minori K. Rosales, Se Hoon Park, Sun Jin Sym, Jichao Sun, Sunnie Kim, Thierry Alcindor, Yee Chao, Yeul Hong Kim, Yoon-Koo Kang, Cheng Ean Chee, Hope E. Uronis, Jennifer Yen, Justin I. Odegaard, Errin Lagow, Matthew C. H. Ng, Kristen A. Marrone, Howard C. Hochster, Paul A. Moore, Jong Gwang Kim, Haeseong Park
Review and revision of manuscript: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
ACKNOWLEDGMENT
Medical writing and/or editorial assistance was provided by Meredith Rogers, MS, and Francesca Balordi, PhD, of The Lockwood Group (Stamford, CT), funded in part by MacroGenics, Inc.
REFERENCES
- Ignatov T, Gorbunow F, Eggemann H, Ortmann O, Ignatov A. Loss of HER2 after HER2-targeted treatment. Breast Cancer Res Treat. 2019;175(2):401-408. doi:10.1007/s10549-019-05173-4
- Katayama A, Miligy IM, Shiino S, et al. Predictors of pathological complete response to neoadjuvant treatment and changes to post-neoadjuvant HER2 status in HER2-positive invasive breast cancer. Mod Pathol. 2021;34(7):1271-1281. doi:10.1038/s41379-021-00738-5
- Catenacci DVT, Moya S, Lomnicki S, et al. Personalized antibodies for gastroesophageal adenocarcinoma (PANGEA): a phase II study evaluating an individualized treatment strategy for metastatic disease. Cancer Discov. 2021;11(2):308-325. doi:10.1158/2159-8290.CD–20-1408
- Maron SB, Chase LM, Lomnicki S, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res. 2019;25(23):7098-7112. doi:10.1158/1078-0432.CCR-19-1704
- Pectasides E, Stachler MD, Derks S, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov. 2018;8(1):37-48. doi:10.1158/2159-8290.CD-17-0395
- Catenacci DVT, Kang Y-K, Park H, et al; CP-MGAH22-5 Study Group. Margetuximab plus pembrolizumab in patients with previously treated, HER2-positive gastro-oesophageal adenocarcinoma (CP-MGAH22-05): a single-arm, phase 1b-2 trial. Lancet Oncol. 2020;21(8):1066-1076. doi:10.1016/S1470-2045(20)30326-0
- Janjigian YY, Maron SB, Chatila WK, et al. First-line pembrolizumab and trastuzumab in HER2-positive oesophageal, gastric, or gastro-oesophageal junction cancer: an open-label, single-arm, phase 2 trial. Lancet Oncol. 2020;21(6):821-831. doi:10.1016/S1470-2045(20)30169-8
- Lee J, Franovic A, Shiotsu Y, et al. Detection of ERBB2 (HER2) gene amplification events in cell-free dna and response to anti-HER2 agents in a large Asian cancer patient cohort. Front Oncol. 2019;9:212. doi:10.3389/fonc.2019.00212
- Liang DH, Ensor JE, Liu Z-B, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155(1):139-149. doi:10.1007/s10549-015-3635-5
- Nakamura Y, Okamoto W, Kato T, et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nat Med. 2021;27(11):1899-1903. doi:10.1038/s41591-021-01553-w
- Wang H, Li B, Liu Z, et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur J Cancer. 2018;88:92-100. doi:10.1016/j.ejca.2017.10.032
- Siravegna G, Lazzari L, Crisafulli G, et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell. 2018;34(1):148-162.e7. doi:10.1016/j.ccell.2018.06.004
- Zhang C, Chen Z, Chong X, et al. Clinical implications of plasma ctDNA features and dynamics in gastric cancer treated with HER2-targeted therapies. Clin Transl Med. 2020;10(8):e254. doi:10.1002/ctm2.254
- Maron SB, Chatila WK, Millang BM, et al. Pembrolizumab with trastuzumab and chemotherapy (PTC) in HER2-positive metastatic esophagogastric cancer (mEG): plasma and tumor-based biomarker analysis. J Clin Oncol. 2020;38 (15 Suppl):abstr 4559. doi:10.1200/JCO.2020.38.15_suppl.4559
- Modi S, Andre F, Krop IE, et al. Trastuzumab deruxtecan for HER2-positive metastatic breast cancer: DESTINY-Breast01 subgroup analysis. J Clin Oncol. 2020;38(15 Suppl):abstr 1036. doi:10.1200/JCO.2020.38.15_suppl.1036
- Thompson JC, Carpenter EL, Silva BA, et al. Serial monitoring of circulating tumor DNA by next-generation gene sequencing as a biomarker of response and survival in patients with advanced NSCLC receiving pembrolizumab-based therapy. JCO Precis Oncol. 2021;5:PO.20.00321. doi:10.1200/PO.20.00321
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449-1458. doi:10.1038/s41591-018-0101-z
- Raja R, Kuziora M, Brohawn PZ, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24(24):6212-6222. doi:10.1158/1078-0432.CCR-18-0386
- Wang C, Chevalier D, Saluja J, Sandhu J, Lau C, Fakih M. Regorafenib and nivolumab or pembrolizumab combination and circulating tumor DNA response assessment in refractory microsatellite stable colorectal cancer. Oncologist. 2020;25(8):e1188-e1194. doi:10.1634/theoncologist.2020-0161
- Zhang Q, Luo J, Wu S, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10(12):1842-1853. doi:10.1158/2159-8290.CD-20-0047
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646-650.
- Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313(1-2):139-142. doi:10.1016/s0009-8981(01)00665-9
- Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14(10):898-908. doi:10.1158/1541-7786.MCR-16-0044
- Catenacci DVT, Park H, Shim BY, et al. 1379P Margetuximab (M) with retifanlimab (R) in HER2+, PD-L1+ 1st-line unresectable/metastatic gastroesophageal adenocarcinoma (GEA): MAHOGANY cohort A. Ann Oncol. 2021;32(Suppl 5):abstr 1379P. doi:10.1016/j.annonc.2021.08.1488
- Bartley AN, Washington MK, Colasacco C, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35(4):446-464. doi:10.1200/JCO.2016.69.4836
- Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24(15):3539-3549. doi:10.1158/1078-0432.CCR-17-3831
- FDA approves first liquid biopsy next-generation sequencing companion diagnostic test. News release. FDA; August 7, 2020. Updated August 11, 2020. Accessed February 23, 2023. https://bit.ly/3xOvr6Y
- Zhao D, Klempner SJ, Chao J. Progress and challenges in HER2-positive gastroesophageal adenocarcinoma. J Hematol Oncol. 2019;12(1):50. doi:10.1186/s13045-019-0737-2
- Bang Y-J, Van Cutsem E, Feyereislova A, et al; ToGA Trial Investigators. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687-697. doi:10.1016/S0140-6736(10)61121-X. Published correction appears in Lancet. 2010;376(9749):1302.
- Mack PC, Weber Redman M, Moon J, et al. Residual circulating tumor DNA (ctDNA) after two months of therapy to predict progression-free and overall survival in patients treated on S1403 with afatinib +/– cetuximab. J Clin Oncol. 2020;38 (15 Suppl):abstr 9532. doi:10.1200/JCO.2020.38.15_suppl.9532
- Pascual J, Cutts RJ, Kingston B, et al. Assessment of early ctDNA dynamics to predict efficacy of targeted therapies in metastatic breast cancer: results from plasmaMATCH trial. Cancer Res. 2021;81(4 suppl):abstr PS5-02. doi:10.1158/1538-7445.SABCS20-PS5-02
- Tsang Shaw A, Martini J-F, Besse B, et al. Early circulating tumor (ct)DNA dynamics and efficacy of lorlatinib in patients (pts) with advanced ALK-positive non-small cell lung cancer (NSCLC). J Clin Oncol. 2019;37(15 Suppl):abstr 9019. doi:10.1200/JCO.2019.37.15_suppl.9019
- Saeki H, Oki E, Kashiwada T, et al; Kyushu Study Group of Clinical Cancer (KSCC). Re-evaluation of HER2 status in patients with HER2-positive advanced or recurrent gastric cancer refractory to trastuzumab (KSCC1604). Eur J Cancer. 2018;105:41-49. doi:10.1016/j.ejca.2018.09.024
- Seo S, Ryu M-H, Park YS, et al. Loss of HER2 positivity after anti-HER2 chemotherapy in HER2-positive gastric cancer patients: results of the GASTric cancer HER2 reassessment study 3 (GASTHER3). Gastric Cancer. 2019;22(3):527-535. doi:10.1007/s10120-018-0891-1
- Makiyama A, Sukawa Y, Kashiwada T, et al. Randomized, phase II study of trastuzumab beyond progression in patients with HER2-positive advanced gastric or gastroesophageal junction cancer: WJOG7112G (T-ACT study). J Clin Oncol. 2020;38(17):1919-1927. doi:10.1200/JCO.19.03077
- Janjigian YY, Riches JC, Ku GY, et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in HER2-overexpressing esophagogastric (EG) tumors treated with trastuzumab. J Clin Oncol. 2015;33(3 Suppl):abstr 63. doi:10.1200/jco.2015.33.3_suppl.63
- Robichaux JP, Elamin YY, Vijayan RSK, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell. 2019;36(4):444-457.e7. doi:10.1016/j.ccell.2019.09.001. Published correction appears in Cancer Cell. 2020;37(3):420.
- Yi Z, Rong G, Guan Y, et al. Molecular landscape and efficacy of HER2-targeted therapy in patients with HER2-mutated metastatic breast cancer. NPJ Breast Cancer. 2020;6:59. doi:10.1038/s41523-020-00201-9
- Greulich H, Kaplan B, Mertins P, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109(36):14476-14481. doi:10.1073/pnas.1203201109
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202-209. doi:10.1038/nature13480
- Kim ST, Do I-G, Lee J, Sohn I, Kim K-M, Kang WK. The NanoString-based multigene assay as a novel platform to screen EGFR, HER2, and MET in patients with advanced gastric cancer. Clin Transl Oncol. 2015;17(6):462-468. doi:10.1007/s12094-014-1258-7

Late Hepatic Recurrence From Granulosa Cell Tumor: A Case Report
Granulosa cell tumors exhibit late recurrence and rare hepatic metastasis, emphasizing the need for lifelong surveillance in affected patients.