Proteomic Patterns Find Ca in Men With High PSA
CHAPEL HILL, North Carolina-Mass spectroscopy-based screening of serum samples from men with elevated PSA levels can distinguish benign from malignant disease and significantly reduce the need for biopsies, according to David Ornstein, MD, and his colleagues at the Food and Drug Administration (FDA) and National Cancer Institute (NCI). Dr. Ornstein is assistant professor of surgery, Division of Urology, University of North Carolina School of Medicine. [See Figure]
CHAPEL HILL, North CarolinaMass spectroscopy-based screening of serum samples from men with elevated PSA levels can distinguish benign from malignant disease and significantly reduce the need for biopsies, according to David Ornstein, MD, and his colleagues at the Food and Drug Administration (FDA) and National Cancer Institute (NCI). Dr. Ornstein is assistant professor of surgery, Division of Urology, University of North Carolina School of Medicine. [See Figure]
The work was published in the Proceedings of the 94th Annual Meeting of the American Association for Cancer Research (abstract 5736). [The meeting, originally scheduled for Toronto in April, was held instead in Washington, DC, in July, owing to the outbreak of severe acute respiratory syndrome (SARS).]
The underlying data reflect a further development of surface enhanced laser desorption time-of-flight (SELDI-TOF) mass spectroscopy, a technology that combines sophisticated computer algorithms with protein binding chips in order to differentiate proteomic patterns that indicate the presence or absence of disease.
This approach exploits the fact that diseased organs alter the proteome (defined as the total array of proteins found in serum) by addition, subtraction, or modification of the constituent proteins; moreover, these changes are dynamic, altering as the disease progresses. Only microliters of sample are required, far less than traditional gel electrophoresis approaches.
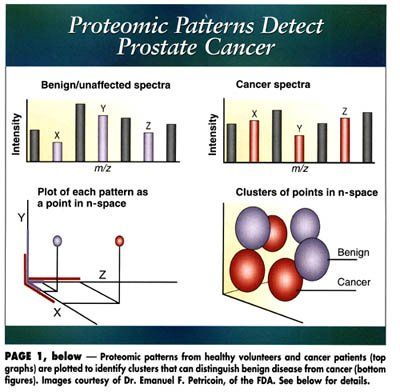
After application of the sample to the chip, a subset of proteins will bind, depending on the particular chromatographic matrix employed; these proteins are then desorbed by laser irradiation, and separated on the basis of their mass-to-charge ratio, all in a matter of minutes. A typical profile is shown in the Figure below, accompanied by the view that would be seen in traditional gel electrophoresis approaches.
By altering the characteristics of the protein chip or the chromatographic matrix, multiple proteomic profiles can be generated from the same serum sample. In order for the technique to be a useful diagnostic tool, it is only necessary to identify specific profiles that reproducibly differentiate the normal and diseased states.
It is important to note that only a correlation is required, meaning that there is no need to identify the proteins whose composition is altered, or to establish any functional link between the changes and the disease state.
The approach does require powerful computational resources and pattern recognition algorithms, however, and imposes strong requirements as to sample reproducibility, but these constraints have proved tractable. In practice, the system must first be "trained" by cataloguing samples from healthy volunteers and from patients who have a given cancer, in order to identify particular profiles of diagnostic interest.
A schematic representation of the technique is shown on page 1. Typically, 5 to 20 proteins will be involved in discriminating between the protein signatures of the population of cancer patients and healthy individuals. Each globular cluster represents the scatter among the patient population for a given proteomic pattern whose presence or absence has diagnostic relevance.
Individual proteomic signatures from new patients, which will consist of a single point in the n-dimensional space, are then compared with these established clusters to make a positive or negative diagnosis. In the event that the patient’s profile does not fit into any of the existing clusters, the program will not be able to render an assessment.
Prostate Cancer Study
The technique has already been employed to diagnose ovarian cancer (Petricoin et al: Lancet 359:572-755, 2002) and prostate cancer (Petricoin et al: J Natl Cancer Inst 94:1576-1578, 2002). In their most recent work, Dr. Ornstein and his colleagues concentrated on men with elevated PSA levels (2.5 to 15 ng/mL), with the aim of reducing the number of follow-up biopsies.
The system was first trained, using serum from 14 men with 2 or more negative biopsies, and 26 men with biopsy-detected prostate cancer; it was then applied in a blinded fashion to samples from 136 men, 35 with prostate cancer, 83 with one negative biopsy, and 18 with two negative biopsies. Sensitivity of detection was 97.1% (34 of 35 cancers detected) while specificity was 55% overall.
"These results are very encouraging," Dr. Ornstein told ONI. "If we had used our algorithm to determine the need for prostate biopsy among these men with an ‘indeterminant’ PSA level, we could have prevented more than half of the unnecessary biopsies and would have only missed one cancer. These results compare favorably to current methods that are being used to improve the specificity of PSA testing such as percent free PSA and complex PSA measurement."
An important advantage of the analytic approach is its adaptive nature, he said; that is, the algorithm is continuously refined as more data are added.
"Prostate cancer is a heterogeneous disease, and although prostate cancer is the second leading cause of cancer deaths among US men, many men are diagnosed with indolent disease and don’t need aggressive treatment. Our hope is to use this new technology to obtain accurate prognostic information that can be used to better advise our patients as to the most appropriate treatment for them," Dr. Ornstein said.
Overall, he said, given further probable technological improvements, he believes that the proteomic screens will be available at costs equivalent to other laboratory tests, "and will offer a degree of precision in diagnosis that is superior to current methodologies," he said.
Prolaris in Practice: Guiding ADT Benefits, Clinical Application, and Expert Insights From ACRO 2025
April 15th 2025Steven E. Finkelstein, MD, DABR, FACRO discuses how Prolaris distinguishes itself from other genomic biomarker platforms by providing uniquely actionable clinical information that quantifies the absolute benefit of androgen deprivation therapy when added to radiation therapy, offering clinicians a more precise tool for personalizing prostate cancer treatment strategies.