Costs and Effectiveness of Genomic Testing in the Management of Colorectal Cancer
Numerous genomic tests are available for use in colorectal cancer, with a widely variable evidence base for their effectiveness and cost-effectiveness. In this review, we highlight many of these tests, with a focus on their proposed role, the evidence base to support that role, and the associated costs and risks.
Table 1: Determining the Cost-Effectiveness of Pharmacogenomic Strategies for Cancer Therapies
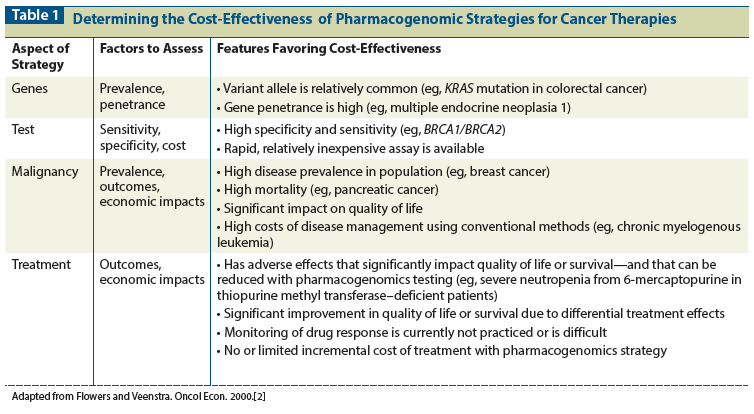
Table 2: Costs and Clinical Roles of Tests
Numerous genomic tests are available for use in colorectal cancer, with a widely variable evidence base for their effectiveness and cost-effectiveness. In this review, we highlight many of these tests, with a focus on their proposed role, the evidence base to support that role, and the associated costs and risks. The tests with the strongest evidence base are KRAS genetic testing in the metastatic setting and microsatellite instability testing in selected patients and in stage II disease. There also may be a role for delineating recurrence-risk signatures for selected patients with stage II disease. The evidence to support broad utilization of uridine 5'-diphospho-glucuronosyltransferase 1A1 (UGT1A1) and dihydropyrimidine dehydrogenase (DPD) testing to guide irinotecan and fluorouracil dosing remains limited. There is much anticipation that next-generation sequencing will herald a new era of targeted therapy for patients with colorectal cancer; however, currently there are no data to support the introduction of widespread testing.
Introduction
Over the past several years, and with the movement toward personalized medicine, there has been significant interest in pharmacogenomics in oncology. A number of tests have been developed that, if they are used appropriately, have the potential to both improve outcomes and decrease costs of care. In this manuscript, we will review the strategies currently available in colorectal cancer and discuss how cost-effectiveness analyses can be applied to evaluate their benefits.
Genomic tests used in oncology predominantly fit into two categories: tumor- and host-specific markers. Tests for tumor-specific markers help to guide the selection of chemotherapy. Host-specific markers are generally used to evaluate a patient’s ability to metabolize certain chemotherapy agents, and thus predict the likelihood of both response and toxicity. In the discussion of different genetic tests, it also is important to recognize the difference between those that assess predictive biomarkers and those that assess prognostic biomarkers.[1] A predictive test provides information related to the risks and benefits of a particular therapy, whereas a prognostic test provides information on the likelihood of survival for patients regardless of the planned treatment.
Thirteen years ago, when the field of pharmacogenomics was in its infancy, Flowers and Veenstra evaluated several pharmacogenomic techniques that were in development and provided a framework for assessing the cost-effectiveness of a pharmacogenomic strategy.[2,3] Since their initial paper, the number of tests available has increased greatly, but the authors’ method of analysis still provides a useful metric for assessing the value of pharmacogenomic interventions. Table 1 illustrates the key factors to consider when assessing the costs and benefits of a pharmacogenomic test for cancer patients.
In the current economic climate and with healthcare costs rising, increasing focus is being placed on analysis of the value of management strategies. While scientific discovery and medical advances are important, they must be balanced against the need for cost containment. The cost-effectiveness of a prognostic test can easily be measured only if it specifically guides treatment decisions. If it is merely informative and not treatment-changing, it has some value to both the patient and clinician, but this is difficult to measure. Prior to performing genomic testing for populations of cancer patients, therefore, there must be evidence that the test is both effective and cost-effective. In some situations, for example, predictive tests may identify a certain patient group that is more likely to benefit from a particular therapy. Such tests can form the basis for a more cost-effective strategy than providing the same treatment to all patients and therefore including a subset who are likely to experience little or no benefit.
In order to evaluate a genomic test, a cost-effectiveness study can be performed. Cost-effectiveness studies are usually performed by using efficacy data from published clinical trials and creating a Markov model with merged financial data, usually from Medicare sources.[4] Efficacy data are commonly adjusted for quality of life, using published utility data, in order to compute quality-adjusted life years (QALYs) gained. The financial costs of toxicities are incorporated into this model. The final results of the analysis are presented as an incremental cost-effectiveness ratio (ICER) for a specific treatment, expressed as a dollar amount for each QALY gained.
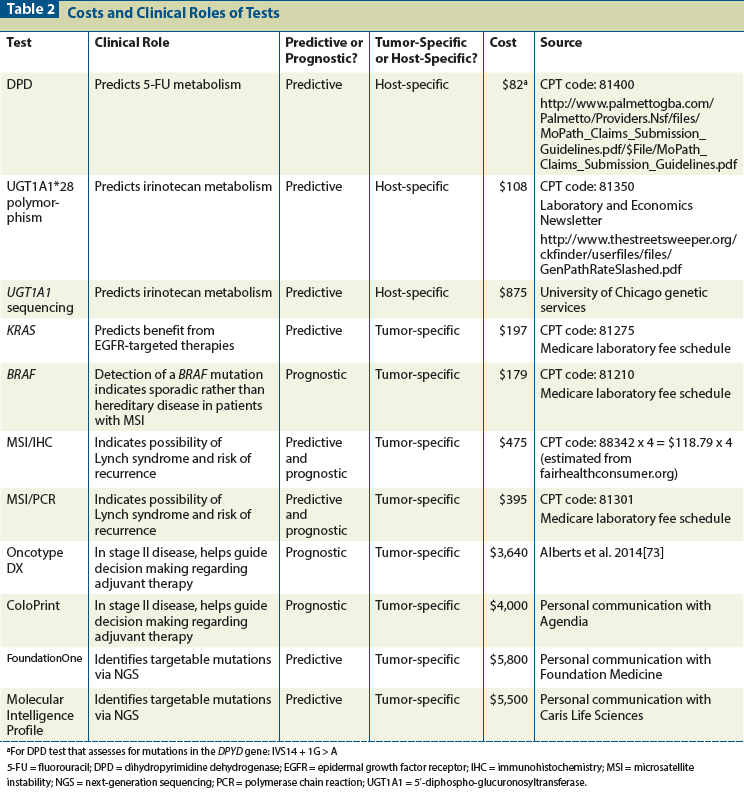
Because the costs of treatment vary considerably both geographically and among payers, sensitivity analyses are commonly performed, using ranges for each parameter within the model. Table 2 identifies several pharmacogenomic tests that have been evaluated for use in the management of colorectal cancer and provides data on costs for specific tests from available sources. To illustrate how these costs relate to the benefits of testing, we discuss specific tests below.
Host-Specific Tests
Dihydropyrimidine dehydrogenase
Fluorouracil (5-FU) is a well-established drug in the management of colorectal cancer. However, it is associated with significant toxicities, such as myelosuppression, hand-foot syndrome, mucositis, and diarrhea. It is metabolized by the enzyme dihydropyrimidine dehydrogenase (DPD), and there has been significant interest in evaluating patient variability in metabolizing 5-FU.[5] The activity of DPD is variable due to genetic variations in the DPYD gene, which encodes the DPD enzyme. Genetic testing was developed with the hope of identifying patients who would be at high risk for severe toxicity when receiving fluoropyrimidine-based chemotherapy. Van Staveren et al recently provided a comprehensive review of numerous methods used to test for DPD deficiency.[5] These authors broadly divided the tests into those that measure enzyme activity and those that detect variants in genes that encode the enzyme.
Enzyme activity can be analyzed in peripheral blood cells. Although this is a highly sensitive and specific technique, it is also very labor intensive and expensive.[6] Another strategy is to assess levels of 2-13C-uracil or its metabolite dihydrouracil, in order to gauge the level of DPD activity. This is accomplished through patient ingestion of 2-13C-uracil, which is subsequently degraded by DPD. The latter strategy is not widely used, due to the limited availability and high cost of both 2-13C-uracil and the spectrophotometer required for its analysis.[5]
Genetic mutations in DPYD can be analyzed by high-performance liquid chromatography. This highly sensitive method screens for heterozygous variants, but it is laborious and time-consuming[5]; in addition, it will detect missense mutations that often have no effect on enzyme activity.[7] Pyrosequencing uses adenosine triphosphate (ATP) to produce light, resulting in the release of a pyrophosphate molecule and the sequential incorporation of bases to the DNA template. This technique for detecting genetic alterations was used to analyze 14 patients who experienced severe 5-FU toxicity; a molecular basis for the deficient phenotype was identified in only 57% of patients.[8] Furthermore, the initial investment for the technology in 2013 was estimated to be between €50,000 and €80,000.[5]
So far, among the many strategies developed for assessing DPD activity, no test has proven to be adequate to mandate testing prior to starting 5-FU–based treatment, due to lack of both effectiveness and cost-effectiveness.[9]
Another technique for individualizing the administration of 5-FU is to use pharmacokinetically (PK)-guided dosing. A phase III trial using PK-guided 5-FU with leucovorin demonstrated improved responses and decreased toxicity.[10] A phase II study demonstrated similar results when PK-guided 5-FU was used with leucovorin and oxaliplatin (FOLFOX).[11] A cost-effectiveness analysis based on this PK-FOLFOX regimen demonstrated it to be cost-effective, with an ICER of $23,000 per QALY.[12]
UGT1A1 testing
UGT1A is an enzyme of the glucuronidation pathway that is encoded by the UGT1A1 gene. It is responsible for the metabolism of steroids, bilirubin, hormones, and drugs such as irinotecan. There are currently 130 documented UGT1A1 mutations,[13] which vary significantly among different ethnic groups.[14] One example of a metabolic disturbance caused by UGT1A1 mutations is Gilbert’s syndrome, a hereditary cause of hyperbilirubinemia that is found in 5% to 10% of the population; it is due to a 70% to 80% reduction in the activity of the UGT1A enzyme secondary to polymorphisms of the UGT1A1 gene.[15]
When irinotecan was in development, a French group reported two cases of patients with Gilbert’s syndrome who received irinotecan and then developed grade 4 neutropenia.[16] Following approval of the drug by the United States Food and Drug Administration (FDA) in 1998, a series of retrospective and prospective studies linked UGT1A1 polymorphism to irinotecan toxicity.[17-21]
A prospective trial in 2004 genotyped 66 patients receiving irinotecan at a dose of 350 mg/m2.[22] Grade 4 neutropenia was found to be highly associated with UGT1A1*28. This led to a recommendation by the FDA for a dose reduction in patients who were homozygous for UGT1A1*28. A subsequent study found that the increased risk of neutropenia was only for the first cycle and did not carry through to subsequent cycles.[23] Improved tumor responses without impact on overall survival (OS) were also noted. Additional genotypes have also been found to be associated with severe toxicity: UGT1A1*60-GG, UGT1A7*3/*3, UGT1A1*93, and UGT1A9*22.[24]
The majority of the studies referenced above were performed to identify patients at increased risk for irinotecan toxicity. However, a corollary to those findings was that consideration could be given to employing an increased dose of irinotecan in patients other than those with UGT1A1*28. A phase 1 dose-escalation study of 59 patients was performed that excluded those with the UGT1A1*28/*28 genotype.[25] The study established that 370 mg/m2 (in *1/*1 patients) and 310 mg/m2 (in *1/*28 patients) can be given safely every 2 weeks.
A meta-analysis did not find statistically significant differences in objective response rate (ORR) between genotype comparison groups.[26] Of all the studies included in the analysis, only the study by Toffoli et al[25] demonstrated statistically significantly improved tumor response for the *28/*28 genotype compared with the wild-type genotype (*1/*1).
A cost-effectiveness analysis of UGT1A1*28 testing was performed.[27] In this modeling study, patients who were homozygous received a 25% dose reduction from the standard dose of 175 mg/m2 to a decreased dose of 131.25 mg/m2. These results showed that 11% of patients would receive a dose reduction, thus avoiding 84.5 cases of severe neutropenia per 10,000 patients and saving $2.7 million in treatment costs. In addition to reducing toxicity, this resulted in an average saving of $272 per patient. Overall, the study showed that pretreatment testing reduced costs.
In another cost-effectiveness study, researchers administered prophylactic granulocyte colony-stimulating factor (G-CSF) to patients who were homozygous for the UGT1A1*28 genotype.[28] The results of this study were severely limited, however, by the lack of inclusion of the cost of G-CSF. Another cost-effectiveness study included the use of G-CSF and dose reduction for patients who were homozygous for UGT1A1*28.[29] The authors concluded that dose reduction was cost-saving in Africans and Caucasians but not in Asians, and that G-CSF use was not cost-saving in any population.
In 2009, the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group produced a recommendation statement regarding UGT1A1 genotyping.[30] This group concluded that the evidence was insufficient to recommend for or against UGT1A1 genotyping prior to irinotecan treatment. The insurance companies Aetna and Cigna currently do not reimburse for the cost of testing, while the National Institute for Clinical Excellence (NICE; United Kingdom), Blue Cross, and Centers for Medicare and Medicaid Services (CMS; United States) do not have a policy regarding testing and instead decide reimbursement requests on a case-by-case basis.[31]
The primary question to be addressed is whether genetic testing prior to the administration of irinotecan therapy is an effective strategy. The prevailing strategy is to institute irinotecan dose reductions for patients who are homozygous for UGT1A1*28; but what should be done if a patient is either wild-type or heterozygous? Evidence suggests that these patients may be receiving suboptimal doses and could tolerate higher doses, possibly with improved responses.[25] Unfortunately, no data exist regarding whether increasing irinotecan doses based on genotype leads to improved survival; a prospective trial is needed to address this question. With the currently available data, we do not recommend testing patients for UGT1A1 polymorphisms to determine the irinotecan dose. We recommend using a reduced dose for patients who develop significant toxicities based on clinical parameters. UGT1A1 polymorphism testing at present remains a research tool that warrants additional studies to determine its effectiveness.
Tumor-Specific Tests
KRAS testing
The introduction into clinical practice of the anti–epidermal growth factor receptor (EGFR) antibodies cetuximab and panitumumab has improved clinical outcomes for patients with metastatic colorectal cancer.[32] It has been noted that KRAS mutations are negative predictive markers for response to these agents.[33] Activating KRAS mutations in codon 12 are detected in approximately 35% to 45% of patients with colorectal cancer[34]; these are a leading cause of resistance to EGFR inhibitors. Mutations in codon 13 of the KRAS gene have been identified and occur in about 6% of patients with colorectal cancer. However, the role of codon 13 mutations in the development of resistance to EGFR treatment remains a matter of controversy. At this point, in the absence of prospective trials and given the contradictory results of the two retrospective studies, the role of KRAS codon 13 mutation in resistance to EGFR inhibition requires further study.[35,36]
There is evidence that NRAS mutations and KRAS mutations in exons 3 and 4 are also negative predictive markers of response. The PRIME trial was a randomized phase III investigation of panitumumab with infusional FOLFOX vs FOLFOX alone as first-line treatment in patients with metastatic colorectal cancer.[37] When patients with any RAS mutation were excluded in the PRIME study, improvement in OS was observed in the panitumumab arm compared with those receiving chemotherapy alone (26.0 vs 20.2 months; P = .043).[38,39] This finding was supported by similar findings in the PEAK trial, a randomized phase II study of panitumumab plus FOLFOX vs bevacizumab plus FOLFOX in patients with previously untreated, wild-type KRAS exon 2 metastatic colorectal cancer.[40] A trend toward a worse outcome was noted with the addition of panitumumab in both the PRIME and PEAK studies in patients with NRAS and non–exon 2 KRAS mutations, suggesting that this group of patients does not benefit and may, in fact, be potentially harmed by anti-EGFR therapy.[38,41] The PICCOLO study analyzed the efficacy of adding panitumumab to irinotecan in patients with KRAS wild-type 5-FU–resistant advanced disease.[42] Patients with any RAS mutation had worse outcomes than did patients with RAS wild-type cancer. In a study using next-generation sequencing on 299 samples from a KRAS wild-type subset of patients receiving panitumumab, similar conclusions were reached.[43] Among KRAS wild-type patients, a treatment effect for progression-free survival (PFS) favoring panitumumab occurred in patients with NRAS wild-type (hazard ratio [HR] = 0.39) and BRAF wild-type (HR = 0.37) but not in those with NRAS mutant-type (HR = 1.94).
The CRYSTAL trial analyzed the survival benefit of adding cetuximab to irinotecan, 5-FU, and leucovorin (FOLFIRI) as first-line treatment for patients with metastatic colorectal cancer.[33] This study confirmed that the addition of cetuximab significantly improved overall response rate (ORR), PFS, and OS compared with FOLFIRI alone in patients with KRAS wild-type disease. The FIRE-3 study compared bevacizumab with cetuximab when added to FOLFIRI in the first-line treatment setting for metastatic colorectal cancer that was exon-2 KRAS wild-type.[44] A subgroup analysis was performed in patients who had KRAS mutations in exons 3 or 4 or NRAS mutations in exons 2, 3, or 4. This subgroup of 34 patients in the cetuximab arm had an ORR of 38%, compared with 65% for patients with wild-type cancers.
The high cost of these targeted therapies remains a concern. A cost-effectiveness analysis showed that without genetic testing, the addition of cetuximab had an ICER of $2.9 million per life year (LY) gained.[45] This is well beyond the threshold of $50,000 to $100,000 per QALY that is considered to be the benchmark of cost-effectiveness.[46] When using KRAS testing as a biomarker, the ICER improved, but only to $2.8 million per LY.[45] While EGFR-targeted therapies do not appear to reach the criteria that warrant their being described as cost-effective, they are commonly used in clinical practice. Due to the observation that RAS testing improves the cost-effectiveness of EGFR-targeted therapies, we therefore support the use of this approach for patients who have metastatic colorectal cancer. However, since there is no evidence to support the use of EGFR-targeted therapy in the adjuvant setting, there is no indication to test for RAS mutations in patients with early-stage disease.
BRAF testing
BRAF is the main downstream effector of KRAS. Mutations in B-Raf proto-oncogene, serine/threonine kinase (BRAF), are found in less than 10% of patients with colorectal cancer. BRAF mutations are a negative prognostic marker regardless of treatment received, with significant differences in median OS: BRAF-mutant, 8.8 months; KRAS-mutant, 14.4 months; and KRAS wild-type, 20.1 months.[47] CRYSTAL analyzed the efficacy of adding cetuximab to irinotecan in the first line of treatment. In a subset analysis of this trial, the impact of utilizing BRAF mutations as a predictive tool in relation to the efficacy of anti-EGFR therapy was examined.[48,49] Patients with mutated BRAF had low response rates and markedly shorter median PFS (5.1 vs 8.3 months) and OS (9.5 vs 22.0 months) compared with patients with wild-type tumors.[50] BRAF inhibition by vemurafenib has been attempted, but with limited responses.[51,52]
The value of BRAF mutation analysis is limited, since it provides prognostic information but does not provide predictive information. However, it does have additional utility in patients with microsatellite instability (MSI). The presence of a BRAF mutation indicates sporadic disease and excludes a diagnosis of Lynch syndrome.[53] Given that prognostic information does not ultimately change management, we propose that it is only necessary to check for BRAF mutation status in patients who have been found to have MSI in order to determine whether it is sporadic or inherited. Furthermore, BRAF and KRAS mutations are mutually exclusive.[47] Therefore, if a KRAS mutation is detected, BRAF status will be wild-type, and there is no need to check for BRAF. For this reason, many institutions use a reflex BRAF test if KRAS is wild-type.
Microsatellite instability testing
The first genetic mutation to cause MSI was discovered in 1993; it was linked to the development of hereditary colon cancer.[54] Further mutations were subsequently discovered, and extensive work has been done to understand the best method of testing and the clinical implications of mutation.[55] Mismatch repair (MMR) genes are responsible for the correction of nucleotide base mispairings and small insertions or deletions that occur during DNA replication.[56] These genes include MSH2, MLH1, PMS1, PMS2, MSH6, and MLH3. Germline mutations in one of the MMR genes is the underlying defect in hereditary nonpolyposis colon cancer, also known as Lynch syndrome.
Approximately 15% of colorectal cancers demonstrate MSI.[57] MSI is the result of germline mutations causing Lynch syndrome in 3%, and the result of sporadic mutations, with no clinical relevance for other family members, in 12%; the remaining 85% of colorectal cancers do not demonstrsate MSI. The presence of a BRAF mutation indicates sporadic rather than hereditary disease and excludes a diagnosis of Lynch syndrome.[53] Sporadic tumors do not contain MMR gene mutations; instead, they have acquired hypermethylation of the MLH1 gene, which silences gene expression. Cells with MMR deficiency accumulate DNA errors throughout the genome.[58] When comparing colorectal cancer that is microsatellite stable (MSS) with colorectal cancer that demonstrates MSI, MSI tumors are seen to have characteristic clinicopathologic features: they tend to occur in the proximal colon, to have a greater mucinous component, to contain lymphocytic infiltration, and to be poorly differentiated.[37]
Multiple studies have demonstrated that the presence of MSI is a good prognostic marker, being associated with decreased rates of recurrence compared with MSS tumors.[57] Furthermore, there is evidence to suggest that MSI patients have a lack of benefit from 5-FU in the adjuvant setting, although the Quick and Simple and Reliable (QUASAR) study had conflicting results with regard to this endpoint.[57]
The revised Bethesda criteria indicate which patients should be tested for MSI.[59] Tumor tissue can be tested using immunohistochemistry to evaluate for the presence of MMR proteins. This method is advantageous, as very few tumor cells are needed, and most pathology labs are able to perform the test simply and quickly.[60] Alternatively, MSI testing can be performed by a polymerase chain reaction assay. This is a more complex and expensive process requiring more tumor cells; however, because of tissue fixation, it has the benefit that results do not vary between laboratories.
We propose that MSI testing be performed in patients who fulfill the revised Bethesda criteria and in any patient with stage II disease, as results may influence decision making regarding the value of adjuvant chemotherapy.
Gene expression profiling to guide decision making in stage II colorectal cancer
In resected stage II colorectal cancer, 5-FU–based chemotherapy confers a 5% improvement in OS at 8 years of follow-up, compared with observation alone.[61] The current standard of care is to give adjuvant chemotherapy only to patients with clinicopathologic factors indicating high risk, such as T4 disease, perforation, obstruction, or lymphovascular/perineural involvement.[62] However, a number of genomic signatures are now available to provide prognostic information based on molecular features of the tumor.
Oncotype DX. This 12-gene expression assay uses a recurrence score, which was validated retrospectively on 1,436 cancer specimens from the QUASAR study, to predict the risk of recurrence.[63] Patients who were found to be at high risk had a 6% absolute improvement in 3-year recurrence rates with adjuvant therapy, while “low-risk” patients had a 3% absolute improvement. This study also confirmed that T4 disease (high-risk) and MSI (low-risk) were the strongest predictors of recurrence. Therefore, in patients with these features, there is no value to the test. In the remainder of stage II tumors, the 3-year risk of recurrence is between 12% and 22%. Although the test provides only prognostic and not predictive information, it appears to be useful in guiding high-risk patients toward and low-risk patients away from adjuvant therapy. A cost-effectiveness analysis projected that Oncotype DX testing would reduce adjuvant chemotherapy use by 17%, compared with current treatment patterns.[64] With the test priced at $3,200, direct medical costs were expected to decrease by an average of $2,971 per patient. This cost reduction is accompanied by the benefit of adjuvant therapy being individually targeted to the patients most likely to benefit.
ColoPrint. This assay classifies a sample as being either high risk or low risk, based on the expression of 18 genes; it was validated prospectively on 114 samples.[65] Low-risk patients had a 5-year relapse-free survival rate of 90%, while high-risk patients had a rate of 74%. The investigators concluded that two-thirds of patients are at low risk and can safely be spared the toxicity and cost of adjuvant therapy. However, it has been noted that these results may have been confounded by unusually high numbers of tumors that had either BRAF mutations or MSI, both of which confer an improved prognosis.[66] A major limitation of this assay is the need for fresh frozen tissue.
Next-generation sequencing
With the recent decrease in the cost of next-generation sequencing (NGS), multiple academic and private institutions have developed panels to check tumors for mutations that may have clinical significance and lead to a more personalized approach to therapy. Currently, there is a wide range of panels evaluating from 5 to 500 mutations. Some of these mutations will lead to clinically actionable information, but many will not. There is no consensus regarding if, when, and how tumors should be sequenced using this method. Furthermore, opinions range widely, and there is no evidence base to employ when deciding how to use the results.
Perhaps the two best-known panels currently available in the United States are produced by Foundation Medicine and Caris Life Sciences. The FoundationOne panel interrogates the entire coding sequence of 315 cancer-related genes plus select introns from 28 genes that are often rearranged or altered in patients with cancer. Results are reported with suggestions for targeted therapies that may be active against the specific mutations identified. This panel was validated with reference samples of pooled cell lines that modeled key determinants of accuracy, including mutant allele frequency, indel length, and amplitude of copy change; it demonstrated a sensitivity and specificity of over 95%.[67] FoundationOne investigators confirmed the accuracy of the panel using 249 formalin-fixed, paraffin-embedded cancer specimens that had been characterized by established assays. When the panel was applied to 2,221 cases, clinically actionable alterations were found in 76% of tumors.
Caris Life Sciences uses immunohistochemistry, fluorescence in situ hybridization, and polymerase chain reaction, in addition to NGS, in order to detect and interrogate each biomarker. A pilot study was performed in 86 patients with refractory cancer to compare PFS when using molecular profiling to guide therapy with PFS of the previous regimen on which the patient had experienced progression (ie, the patient serving as his or her own control).[68] The results, however, have to be viewed with caution, since a comparison cohort was not used as a control population.
Overall, there is great potential for NGS to radically change the way clinical trials are developed and how we treat patients. However, until studies demonstrate the prognostic and/or predictive effectiveness of NGS approaches in specific colorectal cancer populations considering specific treatments, with appropriate comparison populations managed without NGS for validation, the use of these technologies should be limited.
Conclusions and Future Directions
A vast array of genomic tests are available for use in the management of colorectal cancer, with widely varying evidence in terms of their effectiveness and cost-effectiveness. The strongest evidence supports the role of KRAS testing in the metastatic setting and MSI testing in selected patients. There is currently no indication to test patients for DPD. UGT1A1 testing may ultimately be useful, but currently, insufficient data exist to support routine UGT1A1 testing in standard practice. For both 5-FU and irinotecan dosing, the most appropriate strategy is simply to reduce the dose if patients experience unacceptable toxicity. In the case of 5-FU, there may be a role for PK testing to guide dosing.[10,11] There may be a role for genomic signature testing, but only in selected patients with stage II disease. This strategy is now well established in the treatment of patients with breast cancer, and given that its use can prevent a significant proportion of women from receiving unnecessary chemotherapy, it appears to produce meaningful cost savings.[69-71]
Although this has not yet been fully established, prognostic signature testing may provide similar benefits in treating patients with colorectal cancer. The role of NGS is evolving. This is perhaps one of the most exciting areas of modern oncology, and recently the costs of sequencing have been falling. More research is desperately needed to establish a precise role for this strategy, in order for it to meet the criteria of being both an effective and a cost-effective test.
Although the arrival of NGS has generated much excitement in oncology, in many instances, research hopes have not translated into clinical advances.[72] Much care must be taken when sequencing to ensure that there is an appropriate amount of viable tumor within the tested sample, since a single nucleotide polymorphism in DNA may have no implication for the subsequently transcribed RNA. This is one of many issues that have led researchers to take more interest in analyzing the downstream effects of mutations.[72] Also, advances are being made in the emerging fields of transcriptomics (which analyzes RNA transcripts), proteomics (which analyzes the structure, function, and interactions of the proteins produced by particular genes or tissues), and epigenomics (which analyzes the methylation and histone modification of DNA, and the subsequent changes in gene expression that these processes give rise to). Advances in these disciplines will likely play an increasing role in future clinical research.
Financial Disclosure:Dr. Flowers is an unpaid consultant for Genentech and Millennium/Takeda. He also consults for Celgene, Seattle Genetics, OptumRx, Prescription Solutions, and Clinical Care Options. He reports research support from AbbVie, Acerta, Celgene, Gilead Sciences, Infinity Pharmaceuticals, Janssen, Millennium/Takeda, Onyx Pharmaceuticals, Pharmacyclics, and Spectrum. Dr. Goldstein and Dr. Shaib have no significant financial interest or other relationship with the manufacturers of any products or providers of any service mentioned in this article.
References:
1. Thariani R, Veenstra DL, Carlson JJ, et al. Paying for personalized care: cancer biomarkers and comparative effectiveness. Mol Oncol. 2012;6:260-6.
2. Flowers CR, Veenstra D. Will pharmacogenomics in oncology be cost-effective? Oncol Econ. 2000;1:26-33.
3. Flowers CR, Veenstra D. The role of cost-effectiveness analysis in the era of pharmacogenomics. Pharmacoeconomics. 2004;22:481-93.
4. Tumeh JW, Moore SG, Shapiro R, Flowers CR. Practical approach for using Medicare data to estimate costs for cost-effectiveness analysis. Expert Rev Pharmacoecon Outcomes Res. 2005;5:153-62.
5. van Staveren MC, Guchelaar HJ, van Kuilenburg AB, et al. Evaluation of predictive tests for screening for dihydropyrimidine dehydrogenase deficiency. Pharmacogenomics J. 2013;13:389-95.
6. Mattison LK, Ezzeldin H, Carpenter M, et al. Rapid identification of dihydropyrimidine dehydrogenase deficiency by using a novel 2-13C-uracil breath test. Clin Cancer Res. 2004;10:2652-8.
7. Gross E, Ullrich T, Seck K, et al. Detailed analysis of five mutations in dihydropyrimidine dehydrogenase detected in cancer patients with 5-fluorouracil-related side effects. Hum Mutat. 2003;22:498.
8. van Kuilenburg AB, Haasjes J, Richel DJ, et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: identification of new mutations in the DPD gene. Clin Cancer Res. 2000;6:4705-12.
9. Yen JL, McLeod HL. Should DPD analysis be required prior to prescribing fluoropyrimidines? Eur J Cancer. 2007;43:1011-6.
10. Gamelin E, Delva R, Jacob J, et al. Individual fluorouracil dose adjustment based on pharmacokinetic follow-up compared with conventional dosage: results of a multicenter randomized trial of patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:2099-105.
11. Capitain O, Asevoaia A, Boisdron-Celle M, et al. Individual fluorouracil dose adjustment in FOLFOX based on pharmacokinetic follow-up compared with conventional body-area-surface dosing: a phase II, proof-of-concept study. Clin Colorectal Cancer. 2012;11:263-7.
12. Goldstein DA, Chen Q, Ayer T, et al. Cost effectiveness analysis of pharmacokinetically-guided 5-fluorouracil in FOLFOX chemotherapy for metastatic colorectal cancer. Clin Colorectal Cancer. 2014;13:219-25.
13. Canu G, Minucci A, Zuppi C, Capoluongo E. Gilbert and Crigler Najjar syndromes: an update of the UDP-glucuronosyltransferase 1A1 (UGT1A1) gene mutation database. Blood Cells Mol Dis. 2013;50:273-80.
14. Minami H, Sai K, Saeki M, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497-504.
15. Strassburg CP. Hyperbilirubinemia syndromes (Gilbert-Meulengracht, Crigler-Najjar, Dubin-Johnson, and Rotor syndrome). Best Pract Res Clin Gastroenterol. 2010;24:555-71.
16. Wasserman E, Myara A, Lokiec F, et al. Severe CPT-11 toxicity in patients with Gilbert’s syndrome: two case reports. Ann Oncol. 1997;8:1049-51.
17. Ando Y, Saka H, Ando M, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 2000;60:6921-6.
18. Ando Y, Ueoka H, Sugiyama T, et al. Polymorphisms of UDP-glucuronosyltransferase and pharmacokinetics of irinotecan. Ther Drug Monit. 2002;24:111-6.
19. Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43-7.
20. Font A, Sanchez JM, Taron M, et al. Weekly regimen of irinotecan/docetaxel in previously treated non-small cell lung cancer patients and correlation with uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) polymorphism. Invest New Drugs. 2003;21:435-43.
21. Mathijssen RH, Marsh S, Karlsson MO, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9:3246-53.
22. Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382-8.
23. Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061-8.
24. Cecchin E, Innocenti F, D’Andrea M, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457-65.
25. Toffoli G, Cecchin E, Gasparini G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:866-71.
26. Dias MM, McKinnon RA, Sorich MJ. Impact of the UGT1A1*28 allele on response to irinotecan: a systematic review and meta-analysis. Pharmacogenomics. 2012;13:889-99.
27. Gold HT, Hall MJ, Blinder V, Schackman BR. Cost effectiveness of pharmacogenetic testing for uridine diphosphate glucuronosyltransferase 1A1 before irinotecan administration for metastatic colorectal cancer. Cancer. 2009;115:3858-67.
28. Pichereau S, Le Louarn A, Lecomte T, et al. Cost-effectiveness of UGT1A1*28 genotyping in preventing severe neutropenia following FOLFIRI therapy in colorectal cancer. J Pharm Pharm Sci. 2010;13:615-25.
29. Obradovic M, Mrhar A, Kos M. Cost-effectiveness of UGT1A1 genotyping in second-line, high-dose, once every 3 weeks irinotecan monotherapy treatment of colorectal cancer. Pharmacogenomics. 2008;9:539-49.
30. Evaluation of Genomic Applications in Practice and Prevention Working Group. Recommendations from the EGAPP Working Group: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? Genet Med. 2009;11:15-20.
31. Meckley LM, Neumann PJ. Personalized medicine: factors influencing reimbursement. Health Policy. 2010;94:91-100.
32. Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-17.
33. Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-9.
34. Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535-46.
35. De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812-20.
36. Peeters M, Douillard JY, Van Cutsem E, et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab.
J Clin Oncol. 2013;31:759-65.
37. Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697-705.
38. Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-34.
39. Oliner KS, Douillard J-Y, Siena S, et al. Analysis of KRAS/NRAS and BRAF mutations in the phase III PRIME study of panitumumab (pmab) plus FOLFOX versus FOLFOX as first-line treatment (tx) for metastatic colorectal cancer (mCRC). J Clin Oncol. 2013;31:abstr 3511.
40. Schwartzberg LS, Rivera F, Karthaus M, et al. PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240-7.
41. Schwartzberg LS, Rivera F, Karthaus M, et al. Analysis of KRAS/NRAS mutations in peak: a randomized phase II study of FOLFOX6 plus panitumumab (pmab) or bevacizumab (bev) as first-line treatment (tx) for wild-type (WT) KRAS (exon 2) metastatic colorectal cancer (mCRC). J Clin Oncol. 2013;31(suppl):abstr 3631.
42. Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14:749-59.
43. Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res. 2013;19:1902-12.
44. Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065-75.
45. Behl AS, Goddard KA, Flottemesch TJ, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012;104:1785-95.
46. Winkelmayer WC, Weinstein MC, Mittleman MA, et al. Health economic evaluations: the special case of end-stage renal disease treatment. Med Decis Making. 2002;22:417-30.
47. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-62.
48. Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931-7.
49. Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997-2009.
50. Tveit KM, Guren T, Glimelius B, et al. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol. 2012;30:1755-62.
51. Yang H, Higgins B, Kolinsky K, et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res. 2012;72:779-89.
52. Ong FS, Das K, Wang J, et al. Personalized medicine and pharmacogenetic biomarkers: progress in molecular oncology testing. Expert Rev Mol Diagn. 2012;12:593-602.
53. Domingo E, Niessen RC, Oliveira C, et al. BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene. 2005;24:3995-8.
54. Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027-38.
55. Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073-87 e3.
56. Papadopoulos N, Nicolaides NC, Liu B, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915-7.
57. Sinicrope FA, Sargent DJ. Molecular pathways: microsatellite instability in colorectal cancer: prognostic, predictive, and therapeutic implications. Clin Cancer Res. 2012;18:1506-12.
58. Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558-61.
59. Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-8.
60. Bellizzi AM, Frankel WL. Colorectal cancer due to deficiency in DNA mismatch repair function: a review. Adv Anat Pathol. 2009;16:405-17.
61. Sargent D, Sobrero A, Grothey A, et al. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2009;27:872-7.
62. Benson AB 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408-19.
63. Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611-9.
64. Hornberger J, Lyman GH, Chien R, Meropol NJ. A multigene prognostic assay for selection of adjuvant chemotherapy in patients with T3, stage II colon cancer: impact on quality-adjusted life expectancy and costs. Value Health. 2012;15:1014-21.
65. Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17-24.
66. Chee CE, Meropol NJ. Current status of gene expression profiling to assist decision making in stage II colon cancer. Oncologist. 2014;19:704-11.
67. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023-31.
68. Von Hoff DD, Stephenson JJ Jr., Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877-83.
69. Lamond NW, Skedgel C, Rayson D, et al. Cost-utility of the 21-gene recurrence score assay in node-negative and node-positive breast cancer. Breast Cancer Res Treat. 2012;133:1115-23.
70. Holt S, Bertelli G, Humphreys I, et al. A decision impact, decision conflict and economic assessment of routine Oncotype DX testing of 146 women with node-negative or pNImi, ER-positive breast cancer in the U.K. Br J Cancer. 2013;108:2250-8.
71. Blohmer JU, Rezai M, Kummel S, et al. Using the 21-gene assay to guide adjuvant chemotherapy decision-making in early-stage breast cancer: a cost-effectiveness evaluation in the German setting. J Med Econ. 2013;16:30-40.
72. Brooks JD. Translational genomics: the challenge of developing cancer biomarkers. Genome Res. 2012;22:183-7.
73. Alberts SR, Yu TM, Behrens RJ, et al. Comparative economics of a 12-gene assay for predicting risk of recurrence in stage II colon cancer. Pharmacoeconomics. 2014;32:1231-43.