Comparing GOZILA and COLOMATE: Ongoing Umbrella/Basket Trials Examining Genetic Testing in Gastrointestinal Malignancies
Circulating tumor DNA (ctDNA)-based next- generation sequencing (NGS) assays have advantages over classic tissue-based analyses because of their low invasiveness and availability of repeated sampling. The existence of various companion trials for common alterations that can be potential therapy targets on the 2 platforms can lead to future international collaboration.
Bando is head physician in the Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East.

Nakamura is a staff physician in the Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East in Kashiwa, Japan.

Kotani is a staff physician in the Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East.

Yoshino is director of the Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East.

ABSTRACT
Circulating tumor DNA (ctDNA)-based next-generation sequencing (NGS) assays have advantages over classic tissue-based analyses because of their low invasiveness and availability of repeated sampling. Because of the low incidence of target gene alterations such as HER2 or BRAF V600E in gastrointestinal cancers, very large screening platforms are needed to develop genome-based clinical trials. For those reasons, ctDNA-based screening studies are being actively conducted; among them are the GOZILA (Guardant Originates in Zipangu Liquid biopsy Arrival) study in Japan and the COLOMATE (COlorectal Cancer Liquid BiOpsy Screening Protocol for Molecularly Assigned ThErapy) study in the United States.
Although only patients with metastatic colorectal cancer (mCRC) who had previously received standard chemotherapies are eligible for the COLOMATE study, patients with various types of solid tumors at any line of treatment are eligible for GOZILA. This broad coverage of the eligible population allows a target of 5000 patients. By contrast, effective screening of selected candidate patients for companion trials using rapid turnaround time by ctDNA-based NGS assay is the key for COLOMATE. The companion trials of targeted therapies in mCRC are similar between GOZILA and COLOMATE. Both studies have identified patients eligible for studies by examining ERBB2 (HER2), BRAF V600E, BRAF non-V600E, and FGFR alterations, as well as MET amplification and rechallenge with anti-EGFR antibodies. The existence of various companion trials for common alterations that can be potential therapy targets on the 2 platforms can lead to future international collaboration.
Oncology (Williston Park). 2021;35(7):382-389.
DOI: 10.46883/ONC.2021.3507.0382
Introduction
Colorectal cancer (CRC) is the third most common cancer in men and the second most common cancer in women worldwide as of 2020.1 Although several cytotoxic drugs and targeted therapies improve the survival of patients with unresectable disease, median survival is only 30 months.2 Therefore, additional treatment strategies are needed for patients with advanced disease.
Prospective studies based on genomic targets such as ERBB2 (HER2), BRAF V600E, and microsatellite instability (MSI) have demonstrated clinical benefits for patients with metastatic CRC (mCRC).
Cell-free DNA (cfDNA) refers to extracellular DNA molecules, originating from both normal and tumor cells, that are found in body fluids.3 The tumor-derived fraction of cfDNA is commonly called circulating tumor DNA (ctDNA). Analysis of ctDNA has been shown to have huge clinical utility in detecting genetic alterations with minimal invasiveness in various types of cancers, including CRC.4 Additionally, the rapid progress of next-generation sequencing (NGS) technologies for ctDNA analyses have led to significant reductions in sequencing costs, with improved sensitivity and accuracy.
Although tissue-based genotyping is still the standard method for precision oncology, the emerging technology of ctDNA-based genotyping is gradually replacing tissue-based methods in clinical practice.5 In Japan, polymerase chain reaction–based ctDNA assays for EGFR mutations in non–small cell lung cancer and RAS mutations in CRC have been approved and implemented in clinical practice. Moreover, as NGS-based methods have enabled ctDNA profiling with high sensitivity and accuracy and have confirmed analytical validities, several ctDNA-based NGS
assays have been approved in both the United States and Japan.6,7
ctDNA-based NGS assays have various advantages as screening tools for targeted therapies because of their low invasiveness and the availability of repeated sampling. In addition, ctDNA-based assays can detect real-time and spatial genetic status that considers both clonal evolution and tumor heterogeneity. In this review, we explain 2 studies of ctDNA-based screening platforms: GOZILA (Guardant Originates in Zipangu Liquid biopsy Arrival) in Japan (UMIN000029315) and COLOMATE (COlorectal Cancer Liquid BiOpsy Screening Protocol for Molecularly Assigned ThErapy) in the United States (NCT03765736). We also discuss the differences and similarities between the 2 platforms.
SCRUM-Japan GOZILA
To promote genome-based clinical trials in Japan, a nationwide genome screening
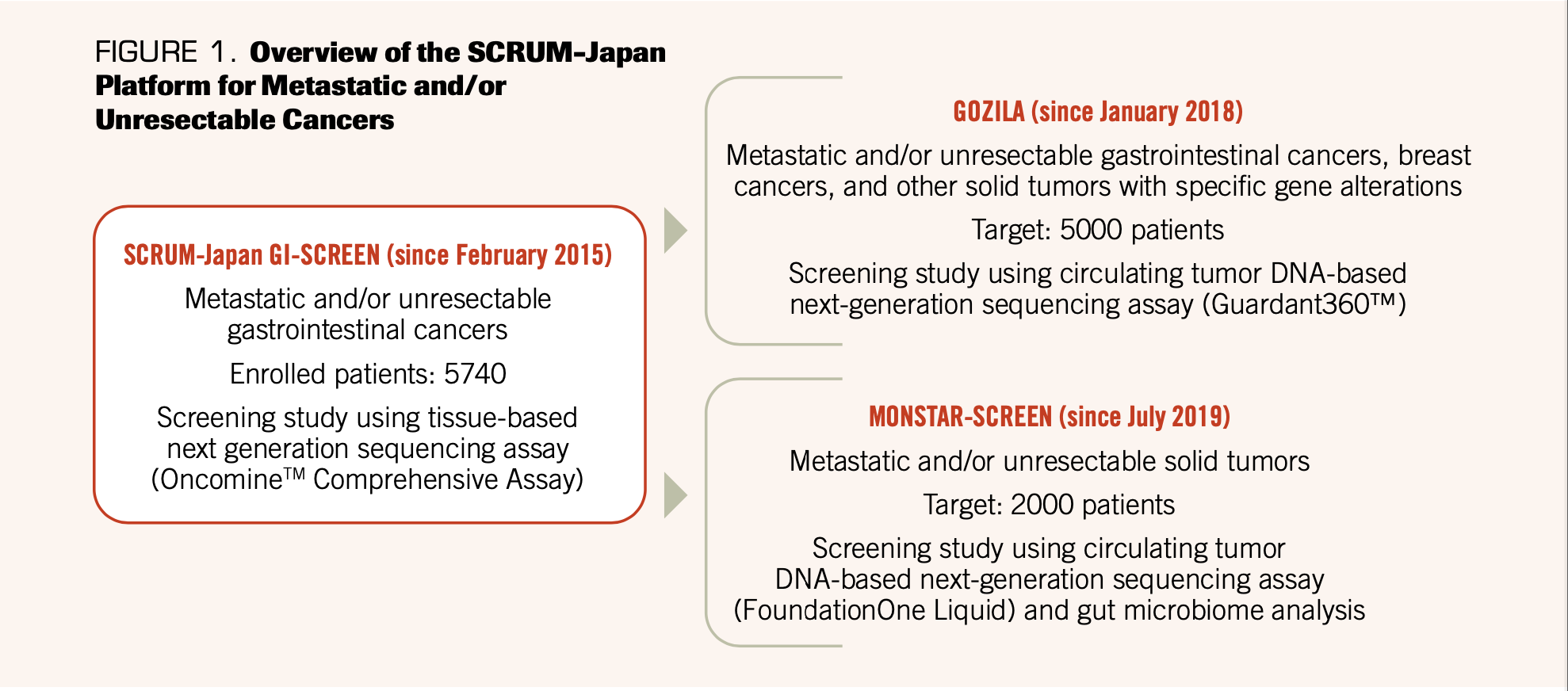
consortium for lung cancer, the LC-SCRUM-Japan, was launched in February 2013. The LC-SCRUM-Japan originally aimed to identify patients harboring ROS1 and RET fusions originally discovered in Japan.8 In February 2014, the GI-SCREEN-Japan multicenter screening project also started for gastrointestinal (GI) cancer. The project began to screen for BRAF, NRAS, and PIK3CA mutations in patients with mCRC.9 In February 2015, these 2 groups merged and initiated the SCRUM-Japan project as a collaboration of industry, government, and academia to achieve nationwide cancer genome screening for precision medicine in Japan (UMIN000016343; UMIN000016344).10 From February 2015 to June 2019, 5740 patients with GI cancer were investigated using the tissue-based Oncomine Comprehensive Assay (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, in January 2018, we initiated GOZILA, a screening study based on the SCRUM-Japan platform using comprehensive ctDNA sequencing by Guardant360™ (Guardant Health, Redwood City, CA, USA) to rapidly screen patients for trial eligibility.11,12 Further, in July 2019, we started MONSTAR-SCREEN (UMIN000036749), a multi-omics profiling and monitoring study using ctDNA sequencing by FoundationOne Liquid (Foundation Medicine, Cambridge, MA, USA) and gut microbiome analysis (Figure 1).
FIGURE 1. Overview of the SCRUM-Japan Platform for Metastatic and/or Unresectable Cancers
Patients are eligible for GOZILA if they have metastatic and/or unresectable GI cancers, including CRC, gastroesophageal adenocarcinoma (GEA), esophageal squamous cell carcinoma (ESCC), pancreatic ductal adenocarcinoma (PDAC), and cholangiocarcinoma (CCA).
Additionally, patients with metastatic and/or unresectable breast cancer and other solid tumors with specific gene alterations diagnosed by quality-assured tissue-based in vitro diagnostic tests are eligible for GOZILA.
Compared with the tissue-based GI-SCREEN project, GOZILA has a significantly shorter sample acquisition duration (median, 4 days vs 14 days; P < .0001) and shorter test duration (median, 7 days vs 19 days; P < .0001). Based on the increased screening speed, GOZILA significantly increased the relative proportion of patients enrolled in clinical trials for targeted therapies (9.5% vs 4.1%; P < .0001).12 However, when we compared the efficacy between GI-SCREEN and GOZILA, patients demonstrated similar objective response rates (20.0% vs 16.7%) and progression-free survival (PFS; median, 2.4 months vs 2.8 months).12
We have also reported the genomic profiles of GI cancers based on ctDNA-based NGS data from the GOZILA study. Patients with ESCC and CRC had the highest ctDNA detection rates (99.1% and 96.0%, respectively), whereas patients with PDAC and GEA had lower detection rates (83.4% and 85.3%, respectively). We also detected multiple biomarkers relevant to therapy selection, including KRAS, NRAS, BRAF, and PIK3CA mutations; ERBB2 (HER2), FGFR1/2, and MET amplifications; FGFR2/3, ALK, NTRK1, and RET fusions; and MSI. In addition, both germline and somatic BRCA mutations were reported in ctDNA samples from patients with PDAC and CCA.12
GOZILA is thus a large and comprehensive ctDNA-based screening platform for GI cancers and other malignancies. GOZILA also significantly shortened the screening turnaround time and improved the trial enrollment rate without compromising treatment efficacy compared with tissue-based screening.
GOZILA-related clinical trials
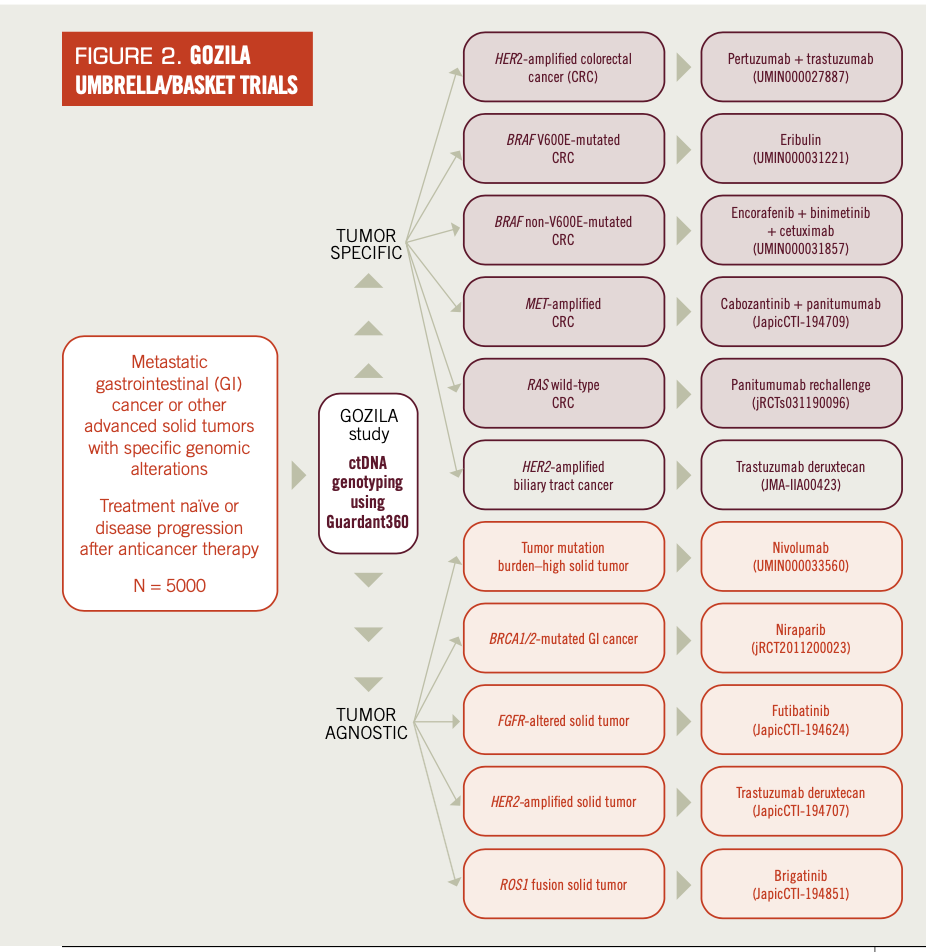
Various types of prospective umbrella/basket clinical trials have been conducted based on this comprehensive GOZILA platform. GOZILA has identified patients for various types of organ-specific umbrella trials for mCRC (Table 1; Figure 2).
TABLE 1. Related Clinical Trials in GOZILA Study
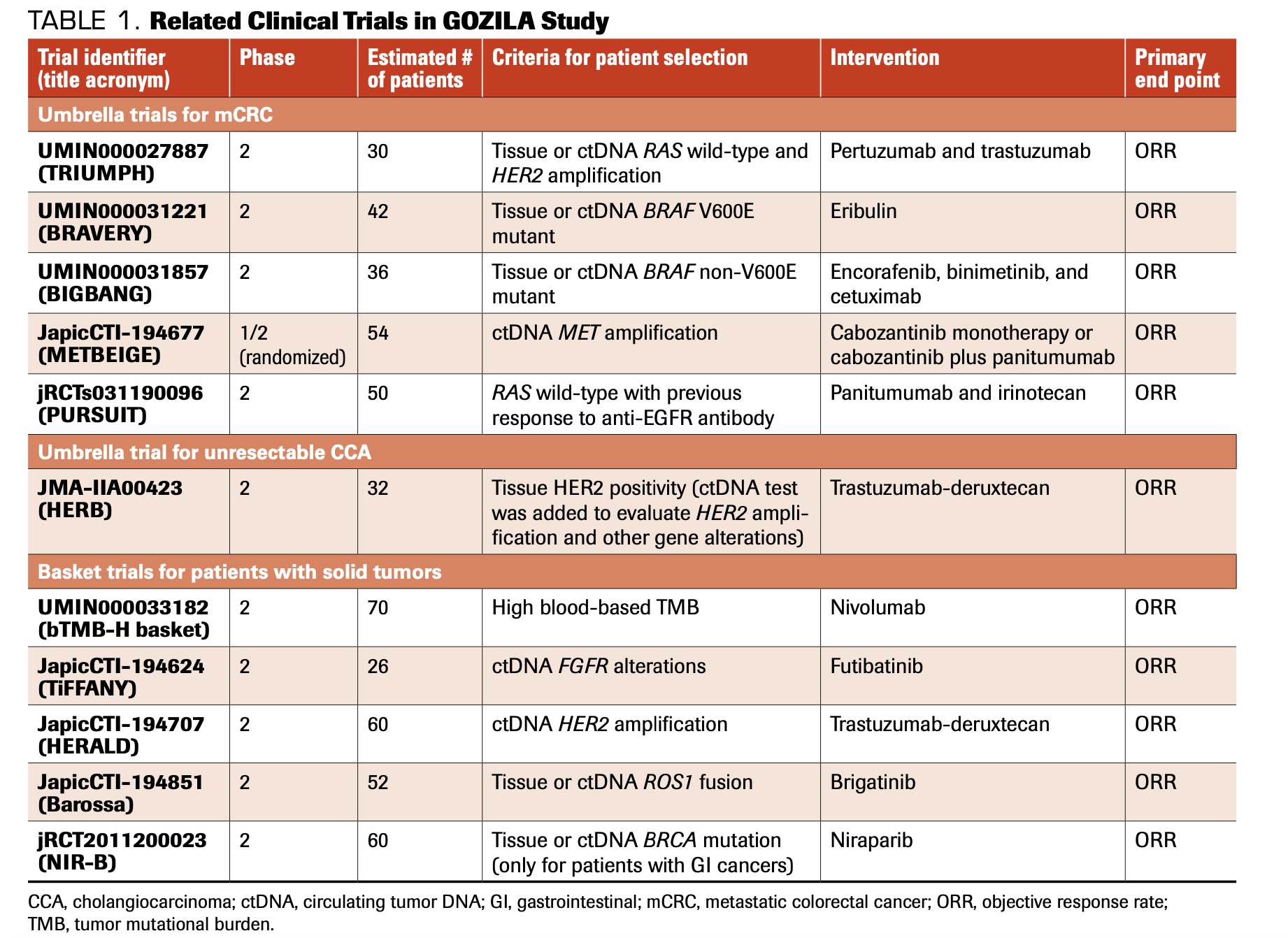
FIGURE 2. GOZILA UMBRELLA/BASKET TRIALS
HER2 amplification occurs in 1% to 4% of patients with mCRC,13-16 and 2 phase 2 trials, HERACLES (NCT03225937) and MyPathway (NCT02091141), showed the efficacy of HER2-targeted therapies.17,18 Therefore, we initiated a single-arm phase 2 trial to evaluate the efficacy of pertuzumab (Perjeta) plus trastuzumab (Herceptin) in patients with mCRC with HER2 amplification that had been confirmed by tumor tissue or ctDNA analysis (TRIUMPH; UMIN000027887). A HER2 positivity rate of 5% to 20% has also been reported among patients with CCA. Trastuzumab deruxtecan (Enhertu) is an antibody-drug conjugate consisting of trastuzumab, a cleavable tetrapeptide-based linker, and a cytotoxic topoisomerase I. Trastuzumab deruxtecan is approved for patients with HER2-positive metastatic gastric and breast cancer in both Japan and the United States. We initiated a single-arm phase 2 trial that investigated the efficacy and safety of trastuzumab deruxtecan in patients with unresectable HER2-positive CCA (HERB; JMA-IIA00423). Although the HER2 status was diagnosed by immunohistochemistry and/or in situ hybridization, the ctDNA test was added to evaluate HER2 amplification and other gene alterations using the GOZILA platform.
BRAF V600E mutation also occurs in 5% to 10% of mCRCs19-22 and is an established factor of poor prognosis. Although the results of BEACON CRC (NCT02928224) indicated that combination therapy with anti-BRAF (encorafenib; Braftovi), anti-MEK (binimetinib; Mektovi), and anti-EGFR (cetuximab; Erbitux) agents led to a significant improvement in survival compared with standard irinotecan-based therapy,23,24 further development of new treatments is very much needed. Because the protein RANBP2 increases microtubule outgrowth from the kinetochores and the protein shRANBP2 impairs proliferation of BRAF V600E–mutant CRC cell lines,25 we conducted a single-arm phase 2 trial to investigate the efficacy and safety of eribulin, a microtubule inhibitor, in patients with BRAF V600E mutations (BRAVERY; UMIN000031221).26 In addition, because BRAF mutations other than V600E (BRAF non-V600E) have been observed in 1.6% to 5.1% of mCRCs27-31 and are increasingly identified by NGS-based assays in patients with mCRC, we initiated a single-arm phase 2 trial to investigate the efficacy and safety of encorafenib, binimetinib, and cetuximab in patients with BRAF non-V600E mutations (BIGBANG; UMIN000031857).32
Further, a longitudinal surveillance of ctDNA by NGS-based assays during anti-EGFR therapy indicated the emergence of acquired RAS mutations and alterations in other genes, including MET, ERBB2 (HER2), FLT3, EGFR, and MEK.33,34 We initiated a randomized phase 2 trial to investigate cabozantinib (Cabometyx) monotherapy vs cabozantinib plus panitumumab (Vectibix) in patients with both primary and acquired MET amplification (METBEIGE; JapicCTI-194709) and a single-arm phase 2 trial to investigate panitumumab plus irinotecan rechallenge in patients with RAS wild-type disease who developed resistance to anti-EGFR antibodies (PURSUIT; jRCTs031190096).
Additionally, GOZILA has several ongoing basket studies to test tissue-agnostic approaches (Figure 2, Table 1).
Tumor mutational burden (TMB) has been established as a predictive biomarker for the efficacy of immune checkpoint inhibitors,35,36 but no prospective clinical trials have investigated the clinical utility of blood-based TMB (bTMB) tests. We are currently investigating the efficacy of nivolumab for treating bTMB-high solid tumors (bTMB-H basket; UMIN000033182). FGFR alterations, including amplifications, mutations, rearrangements, and fusions, are observed in many types of cancer, and pemigatinib (Pemazyre) and infigratinib (Truseltiq) were granted accelerated approval by the FDA for patients with previously treated, unresectable, or metastatic CCA with FGFR2 fusions or other rearrangements. We are currently investigating the efficacy and safety of futibatinib, a pan-FGFR inhibitor, in patients with solid tumors with FGFR alterations (TiFFANY; JapicCTI-194624). In addition to the HERB trial for unresectable CCA, we have investigated the efficacy and safety of trastuzumab deruxtecan for HER2-amplified solid tumors, excluding GEA, breast cancer, lung cancer, CCA, uterine carcinosarcoma, and osteosarcoma, using the ctDNA-based GOZILA platform (HERALD, JapicCTI-194707). Also, for other alterations with tissue-agnostic distributions, we have initiated investigator-initiated studies, including of brigatinib (Alunbrug) for solid tumors with ROS1 fusions (Barossa; JapicCTI-194851) and of niraparib for GI cancers with BRCA mutations (NIR-B; jRCT2011200023).
The results of the TRIUMPH (NCT03413995) and BRAVERY (UMIN000031221) studies have been
reported. In the TRIUMPH study, pertuzumab and trastuzumab were administered every 3 weeks to patients with tissue or ctDNA RAS wild-type and HER2-amplified mCRC.37 The HER2 amplification agreement between the tissue and ctDNA was 83% (62/75). Overall response rate (ORR) was the primary end point, and ORRs of 30% (95% CI, 14%-50%) in 27 tissue-positive patients and of 28% (95% CI, 12%-49%) in 25 ctDNA-positive patients were observed. While 1 partial response (PR), 3 stable disease (SD), and 1 progressive disease (PD) were observed in 5 patients with ctDNA-negative/tissue-positive tumors, only 1 SD and 2 PD were observed in 3 patients with ctDNA-positive/tissue-negative tumors. The confirmed median PFS and median overall survival (OS) were 4.0 months (95% CI, 1.4-5.6) and 10.1 months (95% CI, 4.5-16.5) in tissue-positive patients, respectively, and 3.1 months (95% CI, 1.4-5.6) and 8.8 months (95% CI, 4.3-12.9) in ctDNA-positive patients, respectively. In exploratory analyses, patients with wild-type RAS, BRAF, PIK3CA, and HER2 were more likely to respond to treatment than were those with a mutation (tissue-positive patients: ORR, 36% vs 0%; ctDNA-positive patients, ORR, 32% vs 0%). Decreased ctDNA fraction and HER2 plasma copy number at 3 weeks corresponded to the therapeutic response. In the BRAVERY study, the efficacy and safety of eribulin monotherapy were investigated in patients with tissue- or ctDNA-based BRAF V600E mutations.38 Among the 27 enrolled patients, the ORR was 0%, the disease control rate (DCR) was 41%, the median PFS was 1.4 months, and the OS was 5.2 months. Patients with RAS/AKT and epithelial mesenchymal transition–related gene alterations in ctDNA had a worse DCR (25% vs 54%) and PFS (HR, 2.78; P = .02). At the time of disease progression, acquired gene alterations, such as KRAS and FGFR1 amplification, were detected.
GOZILA provides the broad screening platform for various umbrella/basket clinical trials for targeted therapies, including those in which the targets are ERBB2 (HER2), BRAF V600E, BRAF non-V600E, and MET amplifications; FGFR alterations; ROS1 fusions; BRCA mutations; and TMB. The TRIUMPH study results suggest promising ORRs, PFS, and OS in patients with ctDNA RAS wild-type and HER2-amplified mCRC. Wild-type RAS, BRAF, PIK3CA, and HER2 were also predictors of response to study treatment.
COLOMATE
COLOMATE is a phase 2 umbrella screening trial sponsored by the Academic and Community Cancer Research United consortium. Actionable genomic alterations in patients with mCRC are investigated using Guardant360™, and the impact of molecularly assigned therapy is assessed according to the individual companion studies. Patients are eligible if they have metastatic and/or unresectable CRC and have progressive disease, are intolerant to treatment, or are contraindicated for treatment with a fluoropyrimidine, oxaliplatin, irinotecan, anti-VEGF monoclonal antibodies, or anti-EGFR monoclonal antibodies if RAS wild-type. If the tissue is known to be positive for HER2 (3+ by immunohistochemistry) or the tumor has HER2 amplification as detected by a Clinical Laboratory Improvement Act–certified assay, prior treatment with anti-EGFR therapy is not required.
Several companion studies are ongoing, including panitumumab rechallenge for patients without acquired RAS/BRAF/EGFR extracellular domain mutations or HER2/MET amplification
(PULSE; NCT03992456); tucatinib (Tukysa) plus trastuzumab for patients with HER2 amplification (MOUNTAINEER; NCT03043313); tepotinib (Tepmetko) plus cetuximab for patients with MET amplification; anti-BRAF treatments for patients with BRAF non-V600E mutation or as rechallenge; and pemigatinib for patients with FGFR alterations (Table 2). Additional companion studies are currently underway.
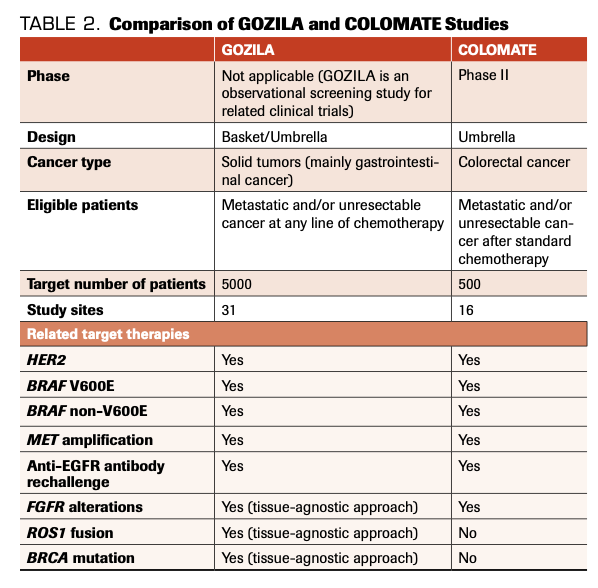
TABLE 2. Comparison of GOZILA and COLOMATE Studies
The primary objectives of COLOMATE are to perform blood-based genomic profiling of patients with treatment-refractory mCRC to facilitate accrual to molecularly assigned therapies and to obtain patient-matched tumor tissue and ctDNA from peripheral blood to facilitate clinically annotated genomic analyses. Secondary correlative objectives are to explore the mechanisms of acquired resistance to molecularly assigned therapy and to explore the correlation between ctDNA-based mutational burden (eg, allele frequency, copy number) and clinical outcomes, such as objective response rate, ORR, PFS, and OS.
MOUNTAINEER is an open-label, pivotal phase 2 trial that initially consisted of a single cohort of up to 45 patients (cohort A) treated with tucatinib plus trastuzumab. Interim analysis of the initial 26 patients enrolled in MOUNTAINEER demonstrated an ORR of 52.2% (12 partial responses in
23 evaluable patients), median duration of response of 10.4 months, median PFS of
8.1 months, and median OS of 18.7 months.39 The trial was expanded to include an additional 70 patients randomized 4:3 into 2 cohorts: Cohort B (N = 40), who will receive tucatinib plus trastuzumab, and Cohort C (N = 30), who will receive tucatinib monotherapy.
Differences and similarities between GOZILA and COLOMATE
In comparing the study designs of GOZILA and COLOMATE, we observed several differences and similarities (Table 2). Only patients with metastatic and/or unresectable CRC are eligible for
COLOMATE, whereas patients with all types of metastatic and/or unresectable GI cancers, breast cancers, and other types of solid tumors with specific gene alterations are eligible for GOZILA. Furthermore, patients at any line of treatment are eligible for GOZILA; as such, its target population is 5000. In contrast, effective screening of candidate patients for companion trials is key for COLOMATE. Using the rapid turnaround time from consent to treatment recommendation by ctDNA-based NGS assay, at least 35% of patients will qualify for a companion trial.
When focusing just on patients with mCRC, the companion trials with targeted therapies are similar between GOZILA and COLOMATE (Table 2). Both groups have identified patients for studies examining HER2 amplification, BRAF V600E mutations, BRAF non-V600E mutations, MET amplification, FGFR alterations, and anti-EGFR antibody rechallenge. Because patients with various types of solid tumors can be eligible for GOZILA, some other basket studies for TMB, FGFR alterations, HER2 amplification, ROS1 fusion, and BRCA mutation are accompanied by tissue-agnostic approaches.
Discussion and conclusions
ctDNA-based genotyping has emerged as a potential tool for evaluating primary and acquired gene alterations in real time. Because the use of ctDNA-based NGS assays is feasible for detecting ctDNA in most patients with GI cancer,12 ctDNA-based approaches can be the future standard platform for basket/umbrella target-based trials. Indeed, companion trials for HER2, BRAF V600E, BRAF non-V600E, and FGFR alterations, as well as MET amplification and anti-EGFR antibody rechallenge, are ongoing in both GOZILA and COLOMATE.
According to the data of ctDNA-based GOZILA and tissue-based GI-SCREEN, ctDNA-based NGS significantly improved sample unavailability (0.3% vs 1.5%) and failure rate (0.1% vs 10.6%). 12 On the other hand, several factors related to lower detection in ctDNA-based analyses have been reported. In GI cancers, lower detection rates of ctDNA were suggested in patients with PDAC and GEA, and in patients with lung-only or peritoneum-only metastatic sites in mCRC.12,40 Lower detections of ctDNA were also reported in patients with early or localized disease41 and in patients undergoing intensive chemotherapy.42 These factors must be taken into consideration when we apply the ctDNA-based genomic analyses.
Although, compared with tissue-based GI-SCREEN, ctDNA-based GOZILA significantly increased the relative proportion of patients enrolled in clinical trials for targeted therapies with similar
efficacies,12 only 9.5% of screened patients were enrolled. To detect further targeted gene alterations and increase enrollment in related clinical trials, we are currently preparing several additional tissue-agnostic studies. Furthermore, whole-exome sequencing (WES) and RNA-based NGS may uncover known fusion events in unexpected tumor histologies and identify novel fusion events with potential clinical relevance.43-45 We are now preparing next-generation comprehensive screening platforms to investigate tissue-based WES and circulating tumor nucleic acid detection, including both ctDNA and ctRNA. Additionally, multi-immunofluorescent staining will be performed to screen a patient’s immune status, including the presence of tumor-infiltrating lymphocytes, macrophages, and immune checkpoint proteins (MONSTAR-SCREEN-2).
In conclusion, both GOZILA and COLOMATE are well-qualified ctDNA-based screening platforms for companion targeted therapies, especially in patients with mCRC. The existence of various companion trials for the same target alteration can lead to future international collaboration with the use of these 2 screening platforms.
*Authors Have equal Contribution
Financial Disclosure: HB reports research funding from AstraZeneca, Sysmex, and honoraria from Taiho Pharmaceutical and Eli Lilly Japan; YN reports research grants from Taiho Pharmaceutical, Chugai Pharmaceutical, Guardant Health, Genomedia, Daiichi-Sankyo, and Seagen; DK reports honoraria from Taiho, Ono, Daiichi-Sankyo, Pfizer, Takeda, Lilly, Merck Biopharma, MSD, Bristol-Myers Squibb, Chugai, and Sysmex; research funding from Ono, MSD, Novartis, IQVIA, Syneos Health, and CMIC ShiftZero; TY reports research grants from Taiho Pharmaceutical, Sumitomo Dainippon, Ono Pharmaceutical, Chugai Pharmaceutical, Amgen, Parexel International, MSD, Daiichi Sankyo, Sanofi.
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi:10.3322/caac.21660
- Yamazaki K, Nagase M, Tamagawa H, et al. Randomized phase III study of bevacizumab plus FOLFIRI and bevacizumab plus mFOLFOX6 as first-line treatment for patients with metastatic colorectal cancer (WJOG4407G). Ann Oncol. 2016;27(8):1539-1546. doi:10.1093/annonc/mdw206
- Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124(2):345-358. doi:10.1038/s41416-020-01047-5
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579-586. doi:10.1200/JCO.2012.45.2011
- Nakamura Y, Shitara K. Development of circulating tumour DNA analysis for gastrointestinal cancers. ESMO Open. 2020;5(Suppl 1):e000600. doi:10.1136/esmoopen-2019-000600
- Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24(15):3539-3549. doi:10.1158/1078-0432.CCR-17-3831
- Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15(9):e0237802. doi:10.1371/journal.pone.0237802
- Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375-377. doi:10.1038/nm.2644
- Bando H, Yoshino T, Shinozaki E, et al. Simultaneous identification of 36 mutations in KRAS codons 61 and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit. BMC Cancer. 2013;13:405. doi:10.1186/1471-2407-13-405
- Bando H. The current status and problems confronted in delivering precision medicine in Japan and Europe. Curr Probl Cancer. 2017;41(3):166-175. doi:10.1016/j.currproblcancer.2017.02.003
- Nakamura Y, Yoshino T. Clinical utility of analyzing circulating tumor DNA in patients with metastatic colorectal cancer. Oncologist. 2018;23(11):1310-1318. doi:10.1634/theoncologist.2017-0621
- Nakamura Y, Taniguchi H, Ikeda M, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26(12):1859-1864. doi:10.1038/s41591-020-1063-5
- Valtorta E, Martino C, Sartore-Bianchi A, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol. 2015;28(11):1481-1491. doi:10.1038/modpathol.2015.98
- Richman SD, Southward K, Chambers P, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol. 2016;238(4):562-570. doi:10.1002/path.4679
- Jeong JH, Kim J, Hong YS, et al. HER2 amplification and cetuximab efficacy in patients with metastatic colorectal cancer harboring wild-type RAS and BRAF. Clin Colorectal Cancer. 2017;16(3):e147-e152. doi:10.1016/j.clcc.2017.01.005
- Sawada K, Nakamura Y, Yamanaka T, et al. Prognostic and predictive value of HER2 amplification in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2018;17(3):198-205. doi:10.1016/j.clcc.2018.05.006
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738-746. doi:10.1016/S1470-2045(16)00150-9
- Meric-Bernstam F, Hurwitz H, Singh Raghav KP, et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019;20(4):518-530. doi:10.1016/S1470-2045(18)30904-5
- De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594-603. doi:10.1016/S1470-2045(10)70209-6
- Nakanishi R, Harada J, Tuul M, et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18(6):1042-1048. doi:10.1007/s10147-012-0501-x
- Tie J, Gibbs P, Lipton L, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer. 2011;128(9):2075-2084. doi:10.1002/ijc.25555
- Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104(5):856-862. doi:10.1038/bjc.2011.19
- Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019;381(17):1632-1643. doi:10.1056/NEJMoa1908075
- Tabernero J, Grothey A, Van Cutsem E, et al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E–mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol. 2021;39(4):273-284. doi:10.1200/JCO.20.02088
- Vecchione L, Gambino V, Raaijmakers J, et al. A vulnerability of a subset of colon cancers with potential clinical utility. Cell. 2016;165(2):317-330. doi:10.1016/j.cell.2016.02.059
- Masuishi T, Taniguchi H, Kotani D, et al. Rationale and design of the BRAVERY study (EPOC1701): a multicentre phase II study of eribulin in patients with BRAF V600E mutant metastatic colorectal cancer. ESMO Open. 2019;4(6):e000590. doi:10.1136/esmoopen-2019-000590
- Ciardiello F, Normanno N, Maiello E, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next-generation sequencing: findings from the CAPRI-GOIM trial. Ann Oncol. 2014;25(9):1756-1761. doi:10.1093/annonc/mdu230
- Haley L, Tseng L-H, Zheng G, et al. Performance characteristics of next-generation sequencing in clinical mutation detection of colorectal cancers. Mod Pathol. 2015;28(10):1390-1399. doi:10.1038/modpathol.2015.86
- Shen Y, Wang J, Han X, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8(12):e81628. doi:10.1371/journal.pone.0081628
- Jones JC, Renfro LA, Al-Shamsi HO, et al. Non-V600 BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol. 2017;35(23):2624-2630. doi:10.1200/JCO.2016.71.4394
- Shinozaki E, Yoshino T, Yamazaki K, et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study. Br J Cancer. 2017;117(10):1450-1458. doi:10.1038/bjc.2017.308
- Kotani D, Bando H, Taniguchi H, et al. BIG BANG study (EPOC1703): multicentre, proof-of-concept, phase II study evaluating the efficacy and safety of combination therapy with binimetinib, encorafenib and cetuximab in patients with BRAF non-V600E mutated metastatic colorectal cancer. ESMO Open. 2020;5(1):e000624. doi:10.1136/esmoopen-2019-000624
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):827. doi:10.1038/nm0715-827b
- Strickler JH, Loree JM, Ahronian LG, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018;8(2):164-173. doi:10.1158/2159-8290.CD-17-1009
- Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10(12):1808-1825. doi:10.1158/2159-8290.CD-20-0522
- Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353-1365. doi:10.1016/S1470-2045(20)30445-9
- Okamoto W, Nakamura Y, Kato T, et al. Pertuzumab plus trastuzumab and real-world standard of care (SOC) for patients (pts) with treatment refractory metastatic colorectal cancer (mCRC) with HER2 (ERBB2) amplification (amp) confirmed by tumor tissue or ctDNA analysis (TRIUMPH, EPOC1602). J Clin Oncol. 2021;39(Suppl 15):abstr 3555. doi:10.1200/JCO.2021.39.15_suppl.3555
- Masuishi T, Taniguchi H, Kotani D, et al. Rationale and design of the BRAVERY study (EPOC1701): a multicentre phase II study of eribulin in patients with BRAF V600E mutant metastatic colorectal cancer. ESMO Open. 2019;4(6):e000590. doi:10.1136/esmoopen-2019-000590
- Strickler JH, Zemla T, Ou F-S, et al. Trastuzumab and tucatinib for the treatment of HER2 amplified metastatic colorectal cancer (mCRC): initial results from the MOUNTAINEER trial. Ann Oncol. 2019;30(Suppl 5):v200. doi:10.1093/annonc/mdz246.005
- Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28(6):1325-1332. doi:10.1093/annonc/mdx125
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi:10.1126/scitranslmed.3007094
- Montagut C, Siravegna G, Bardelli A. Liquid biopsies to evaluate early therapeutic response in colorectal cancer. Ann Oncol. 2015;26(8):1525-1527. doi:10.1093/annonc/mdv228
- Singh H, Li YY, Spurr LF, et al. Molecular characterization and therapeutic targeting of colorectal cancers harboring receptor tyrosine kinase fusions. Clin Cancer Res. 2021;27(6):1695-1705. doi:10.1158/1078-0432.CCR-20-4073
- Tsang ES, Grisdale CJ, Pleasance E, et al. Uncovering clinically relevant gene fusions with integrated genomic and transcriptomic profiling of metastatic cancers. Clin Cancer Res. 2021;27(2):522-531. doi:10.1158/1078-0432.CCR-20-1900
- Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25(15):4712-4722. doi:10.1158/1078-0432.CCR-19-0225
