Profilin 1 Protein and Its Implications for Cancers
Profilin 1, or PFN1, is a key actin-binding protein that is involved in various cellular activities, such as cell motility, survival, and membrane trafficking. By summarizing the functions of PFN1 in cancer, investigators hope to better understand the mechanisms of PFN1 in cancer progression.
Wang is from the Department of Respiratory Medicine in the Department of Respiratory and Critical Care Medicine, National Key Clinical Specialty at Xiangya Hospital of Central South University in Hunan, China.

Wang is from the Department of Respiratory Medicine in the Department of Respiratory and Critical Care Medicine, National Key Clinical Specialty at Xiangya Hospital of Central South University in Hunan, China.

Wan is from the Department of Respiratory Medicine in the Department of Respiratory and Critical Care Medicine, National Key Clinical Specialty at Xiangya Hospital of Central South University in Hunan, China.

Hu is from the Department of Respiratory Medicine in the Department of Respiratory and Critical Care Medicine, National Key Clinical Specialty at Xiangya Hospital of Central South University in Hunan, China.

Lu is from the Department
of Oncology in the Hunan Provincial People’s Hospital/ The First Affiliated Hospital of Hunan Normal University in Changsha, Hunan, PR China

Introduction
Profilin 1 (PFN1) is a ubiquitous small-molecule protein that exists in all eukaryotes.1 PFN1 was first identified as a G-actin sequestering molecule,2 and subsequently, its true functions in actin polymerization and F-actin dynamics were revealed.3 In the following decades, the structure of PFN1 was recognized to have 3 domains: an actin-binding domain,4 a poly-L-proline (PLP)-binding domain,5 and a phosphoinositide-binding domain.6
PFN1 plays a vital role in many cell functions, including membrane trafficking, endocytosis, cell cycle, motility, proliferation, cell survival, transcription, stemness, and autophagy (Figure 1). Abnormal expression or deletion of PFN1 can affect the normal physiological activity of cells and lead to disease development. PFN1 has been deeply studied in a variety of diseases, some genetic (eg, amyotrophic lateral sclerosis)7 and some chronic (eg, hypertension).8
FIGURE 1. Functions of Profilin 1 in Cancer
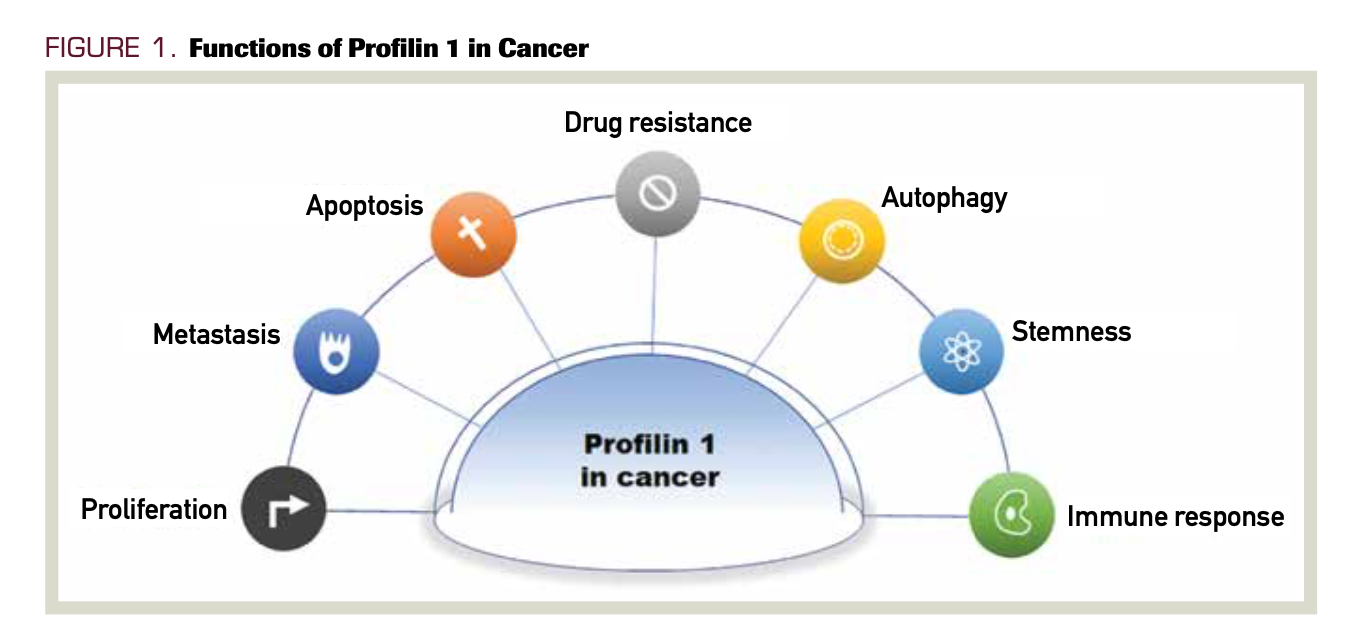
In the past 10 years, PFN1’s role in cancer has received increasing attention. In this review, we summarize the studies of PFN1 in cancer that have been completed in recent years, discuss the roles of PFN1 in cancer, and discuss the implications for tumor diagnosis and therapy in the future.
Oncology (Williston Park). 2021;35(7):402-409.
DOI: 10.46883/ONC.2021.3507.0402
PFN1 as a biomarker for the diagnosis and prognosis
of cancer
Early diagnosis of cancers is still a major challenge worldwide, and early detection can notably reduce their associated morbidity and mortality.9 PFN1, a critical actin-binding protein, is found to be dysregulated in many cancers, which makes it possible to use it as a biomarker for diagnosis and prognosis. PFN1 mainly plays a role in the cytoplasm, but it can also be found in the nucleus and can even be secreted into the extracellular space. The rich knowledge in the proteomics field makes the detection of proteins for new diagnostic markers and targets for therapy possible.10
In some tumor types (such as renal cell carcinoma [RCC], gastric cancer, and others), high expression of PFN1 indicates later stage and worse prognosis. Via differential proteomics, PFN1 has been identified in metastatic and primary RCC, and further analysis indicated that high PFN1 expression was associated with poor outcome and that PFN1 could be used as a potential prognostic marker in RCC.11 In clear-cell RCC (ccRCC), the expression of PFN1 was decreased in early-stage tumors compared with normal tissues. However, its expression in stage IV ccRCC was significantly increased. PFN1 was selected as a candidate marker of late-stage ccRCC.12 Results of a recent study determined that the vast majority of ccRCC tumors tend to be selectively PFN1-positive in stromal cells only; dramatic transcriptional upregulation of PFN1 was found in tumor-associated vascular endothelial cells in clinical specimens of ccRCC.13 Tissue microarray results also showed that PFN1 was increased in metastatic ccRCC compared with primary tumors. Univariate analysis suggested that higher PFN1 expression was associated with shorter disease-free survival (HR, 7.36; P = .047) and lower overall survival.14
In gastric cancer, Tanaka et al found that PFN1 was highly expressed in fetal rat stomach. Additionally, PFN1 was overexpressed in some human and rat gastric cancers.15 The results of later studies indicated that PFN1 expression was higher in gastric cancer tissues than in adjacent normal tissues. High PFN1 expression was correlated with tumor infiltration, lymph node metastasis, and tumor-node-metastases (TNM) stage. Functional assays confirmed that silencing PFN1 could inhibit the invasion and migration of gastric cancer cell lines.16
In addition, PFN1 expression was higher in non–small cell lung cancer (NSCLC). Lower expression of PFN1 was associated with better prognosis and a higher survival rate in NSCLC.17 Proteomic analysis revealed that PFN1 was differentially expressed in laryngeal carcinoma tissues compared with adjacent normal tissues. Further study results revealed that PFN1 was increased in laryngeal carcinoma tissues compared with adjacent normal tissues, indicating that PFN1 was a novel potential biomarker for the diagnosis of laryngeal carcinoma.18
However, in some other tumors (such as colorectal cancer [CRC], oral carcinoma, and others), the opposite is true. PFN1 was downregulated in pancreatic cancer.19-20 Lower expression of PFN1 was significantly associated with a shorter survival period.20 In late-stage oral squamous cell carcinoma, PFN1 expression was lower than that in normal oral epithelium, and loss of PFN1 expression was related to invasion into and metastasis of lymph nodes.21 PFN1 was also decreased in late advanced hepatocellular carcinoma (HCC) and was associated with a poor survival rate of patients.22-23 In addition, PFN1 was found to be downregulated in nasopharyngeal carcinoma24 and breast cancer.25 Combined with another 4 actin-binding proteins, PFN1 could be used to construct a model for predicting poor prognosis of esophageal squamous cell carcinoma.26
Under normal physiological conditions, PFN1 is involved in multiple cellular functions, such as cell motility, migration, adhesion, and transduction signaling pathways.27 PFN1 is differentially expressed in various types of tissues and cells, which may explain its variable tumorigenic mechanisms in different tumors, even in different stages of the same cancer (Figure 2). Because PFN1 plays important roles in tumorigenesis and progression, targeting PFN1 dysregulation could to some extent influence the prognosis of patients with cancer. Determining the expression of PFN1 could thus be used to distinguish high-risk disease from lower-risk disease. Combination with other indices could further improve the diagnostic and prognostic value of PFN1.
FIGURE 2. Two Sides of Profilin 1 in Different Tumor Types
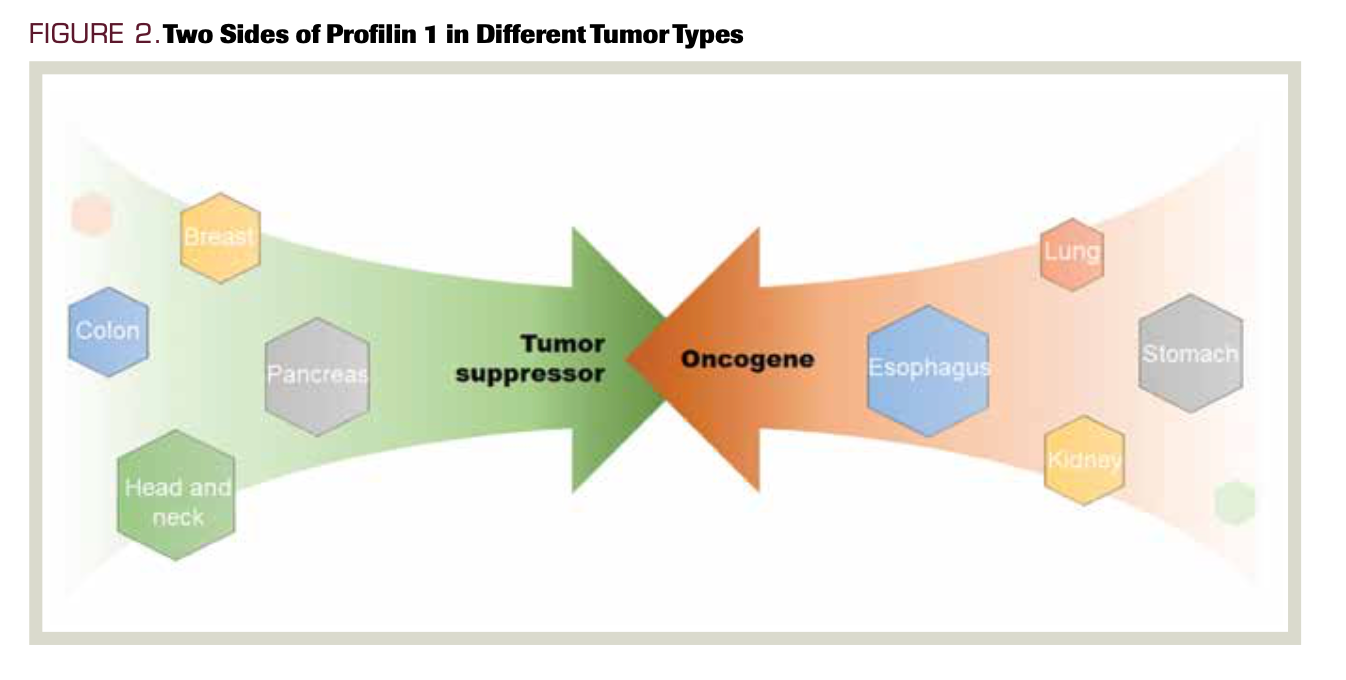
In addition to dysregulation in tumor tissues, PFN1 was also found to be
differentially expressed in the serum, urine, and extracellular vesicles of patients with cancer, which makes it possible to utilize PFN1 in liquid biopsy analysis of tumors. Compared with tumor tissue biopsy, liquid biopsy is a more practical method for real-time monitoring of patients with cancer.28 In addition, PFN1 was detected in the supernatants of cultured cells.
It has been shown that PFN1 gene expression is increased in peripheral blood cells of patients with HCC compared with healthy controls.29 A 9-gene expression system (including PFN1) was used to discriminate patients with HCC from healthy people.30 Proteomic analysis of serum proteins showed that PFN1 was increased in patients with gallbladder cancer. The expression difference between these patients and healthy controls was more than 2-fold.31 PFN1 was differentially expressed in the urine of patients with invasive and noninvasive bladder cancer. Further studies confirmed that PFN1 was notably decreased in the epithelium of invasive bladder tumors compared with noninvasive tumors, which was associated with the clinical outcomes of bladder cancer.32 In in vitro pancreatic cancer cell lines, PFN1 was downregulated in secretomes compared with nonneoplastic pancreatic ductal cells.33 In invitro cultured RCC cell lines, PFN1 was differentially regulated in the supernatant. Further studies revealed that PFN1 was upregulated in RCC tissues.34 Apart from its dysregulation in serum and urine, PFN1 was found to be downregulated in the circulating leukocytes of patients with breast cancer compared with healthy controls, which provides a new paradigm for highly sensitive and less invasive approaches for the diagnosis of breast cancer.35 Studies have already revealed that PFN1 can be secreted via exosomes or other secretory pathways.36-38
Extracellular PFN1 in the tumor microenvironment can be taken up by recipient cells and execute its function in recipient cells, which in turn may influence the biological behavior of cells in the microenvironment, ultimately affecting tumorigenesis and progression of cancers. As mentioned above, PFN1 is expressed differentially in the serum and urine of patients with cancer, which enables its application as a biomarker for diagnosis and prognosis in liquid biopsy (Table 1).
TABLE 1. Aberrant Expression of Profilin 1 in Various Cancers
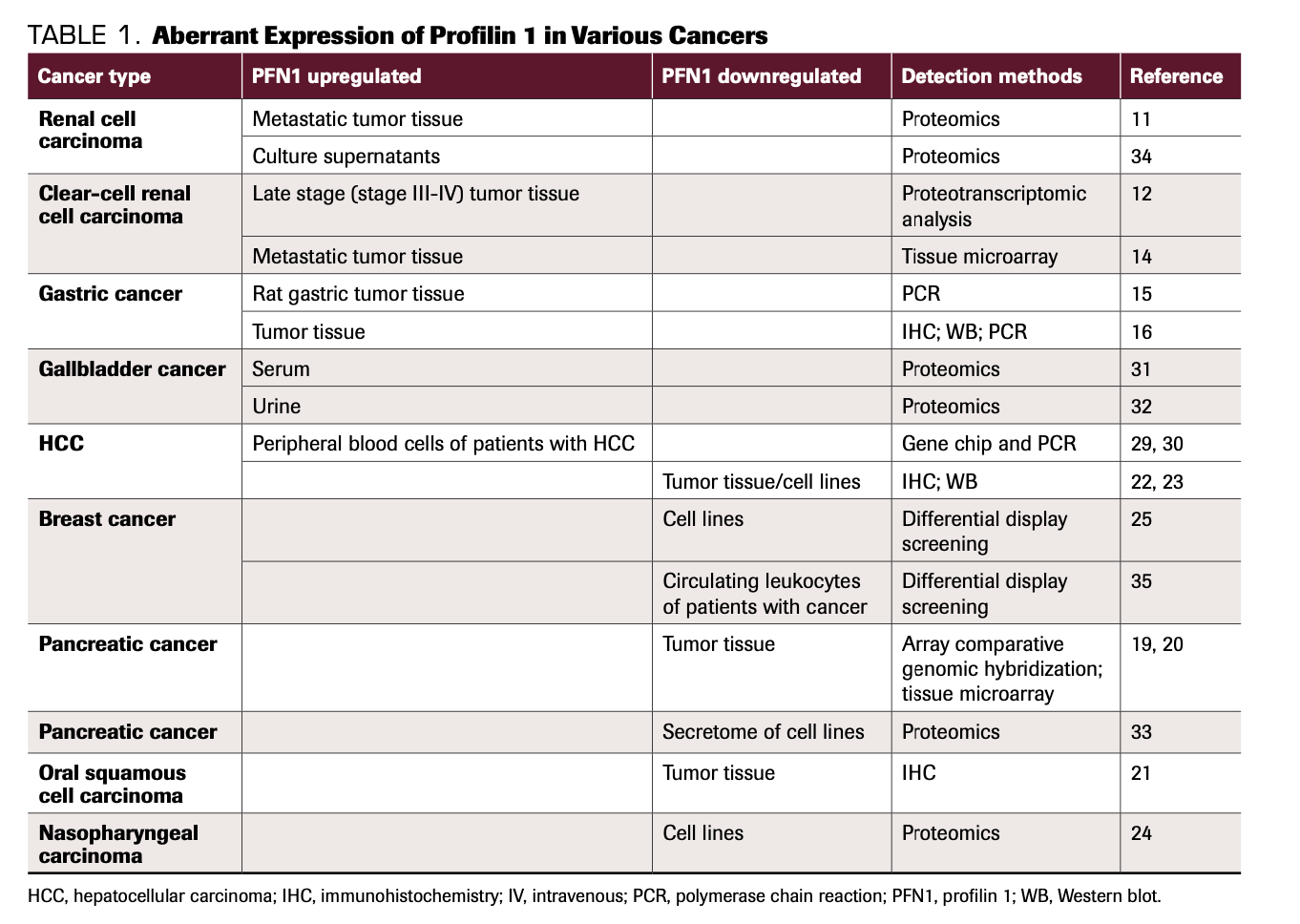
PFN1 as a promoter or inhibitor of cancer metastasis
Cell motility involves membrane protrusion, cell matrix adhesion, cell body translocation, and rear detachment. Many of these processes require the actin cytoskeleton and its regulators. By facilitating the exchange of ATP for ADP on G-actin, PFN1 plays a major role in actin polymerization, thus influencing motility in numerous cells.39 PFN1 also participates in cell motility by regulating actin polymerization and interactions with other regulators of actin cytoskeletons, such as ARP3, VASP, and proteins of cell signaling pathways. Cell-cell adhesion and cell-matrix adhesion are critical contributors to maintaining tissue architecture. Dysregulation of cell-cell adhesion is an important sign in tumor initiation and progression of malignancy. PFN1 can modulate cell adhesion and epithelial-to-mesenchymal transition (EMT) in cancer cells. However, the mechanisms by which PFN1 regulates cell adhesion are still not very clear. Undoubtedly, learning more about the roles of PFN1 in cell adhesion and motility will help us better understand its roles in modulating tumor invasion and migration.
Since PFN1 plays a critical role in actin polymerization, it is an indispensable regulator of cell motility. PFN1 participates in the invasion and metastasis of multiple cancers. However, the roles of PFN1 in regulating cell motility are context specific.27 Exogenous PFN1 with intact actin-binding abilities can ameliorate the adherence and spreading capabilities of cancer cells and exert tumor-suppressive effects in breast cancer.40 Consistent with the results of the study by Wittenmayer et al, Zou et al found that PFN1 overexpression could revert MDA MB-231 cells to an epithelioid phenotype, with restored adherence junctions.41 In addition, PFN1 overexpression could promote AMPK activation and p27 phosphorylation, which in turn induces epithelial morphological reversion of mesenchymal breast cancer through restoration of adherens junctions.42 These studies highlighted the involvement of PFN1 in epithelial adhesion and differentiation, which helped us better understand its roles in cancer cell motility.
Invadopodia are actin-driven membrane protrusions that can deliver matrix metalloproteinases to degrade the matrix and support invasion and dissemination of tumor cells. Any dysregulation of the actin cytoskeleton can impair the formation and maturation of invadopodia.43-46 PFN1 can regulate PI(3,4)P2, which in turn negatively regulates lamellipodin at the leading edge of breast cancer cells and thus inhibits those cells’ motility.47 The depletion of PFN1 leads to an increase in the level of PI(3,4)P2 in invadopodia and its interacting adaptor Tks5. The interaction of PI(3,4)P2-Tks5 has been shown to promote the anchorage, maturation, and turnover of invadopodia, which in turn enhances the invasiveness and motility of breast cancer.48 Breast cancer is an invasive adenocarcinoma, and numerous studies have found that PFN1 is downregulated in breast cancer tissues.49-54 Overexpression of PFN1 reduces the invasion and migration of breast cancer cells, while loss of PFN1 significantly enhances breast cancer cell motility and invasion. Mechanisms involved in PFN1’s negative roles in breast cancer metastasis include Enabled (Ena)/vasodilator stimulated phosphoprotein (VASP)-dependent lamellipodial protrusion,51 miRNA-182 regulation,52 and regulation of PFN1 degradation.53 Mouneimne et al found that PFN1 knockdown (KD) could increase F-actin bundles and enhance stress fiber formation. In that study, the numbers of protrusions in PFN1-KD cells were markedly decreased, and PFN1-KD could inhibit the motility of breast cancer.55 Moreover, Liu et al indicated that the interaction of LMO2-PFN1 and LMO2-ARP3 could promote the formation of lamellipodia/filopodia in basal-type breast cancer cells.56 Ena/VASP is a critical regulator of the actin cytoskeleton at the leading edge of cells, which controls membrane protrusions and cell motility. Cell-substrate adhesion and downregulation of Protein Kinase A (PKA) promote interactions of PFN1 with VASP, which is another mechanism by which PFN1 regulates cell motility.57-58 Knockdown of PFN-1 has been shown to abrogate the inhibitory effect of tyrphostin A9, suggesting that modulating PFN1 expression could have therapeutic potential in the treatment of metastatic breast cancer.59
As in breast cancer, PFN1 was found to be a suppressor of migration in HCC.22,23,60 All-trans retinoic acid60 and guttiferone K22 could inhibit hepatocellular cell migration and proliferation by upregulating the expression of PFN1. In prostate cancer, cathepsin X can inactivate PFN1, thus promoting adhesion, invasion, and migration of cancer cells.61 In CRC, elevated expression of PFN1 obviously inhibited invasion and migration. PFN1 was suppressed by the HLA-F-AS1/miRNA-330-3p/PFN1 or HCP5/miRNA-299-3p/PFN1/AKT axis.62-63
Interestingly, Ding et al showed that in the early stages of metastasis, breast cancer cells exhibit a hyperinvasive phenotype characterized by upregulation of MMP-9 and by faster invasion when PFN1 expression is downregulated. However, in the late stages of metastasis, loss of PFN1 markedly inhibits the growth of metastatic colonies of breast cancer cells.54 Rizwani et al reported that PFN1 expression was elevated in breast cancer tissues and that overexpression of PFN1 could inhibit the migration of breast cancer cells. The phosphorylation of S137 mutants abrogated PFN1’s promotion of migration. These studies provided a different vision of PFN1’s role in breast cancer metastasis.64
In gastric cancer, silencing PFN1 inhibited the invasion and migration of cells, and the PFN1 expression level in cancer tissue was positively correlated with tumor infiltration and lymph node metastasis.16 However, different conclusions were drawn from the study of Ma et al. The authors found that PFN1 expression was inversely correlated with lymph node metastasis.65 In the lung cancer cell line A549, downregulation of PFN1 inhibited migration.17 In addition, in vitro studies support the importance of PFN1 in the proliferation and migration of RCC cells, and treatment with a novel computationally designed PFN1-actin interaction inhibitor reduced the proliferation and migration of RCC cells in vitro and RCC tumor growth in vivo.13 Additional studies have demonstrated that downregulation of PFN1 can also suppress the migration of laryngeal cancer18 and bladder cancer.66
Although more studies on PFN1 have been completed recently, its roles in cancer metastasis are still unclear. The concentrations of actin and PFN1 are time- and space-specific, and so is the regulation of the actin cytoskeleton (Table 2). Additional thorough studies are needed to comprehend the mechanisms and laws regulating the actin cytoskeleton. More importantly, in addition to actin dependence, PFN1 affects cell migration in an actin-independent manner by interacting with proteins with PIP2 or PLP domains. Furthermore, lncRNAs and microRNAs also modulate the functions of PFN1. All of these proteins and RNAs interact with PFN1 and indirectly influence the functions of cancer cells, which makes understanding the roles of PFN1 in cancer metastasis and other functions more complicated (Table 3).
TABLE 2. Mechanisms of PFN1 in Metastasis of Different Cancers
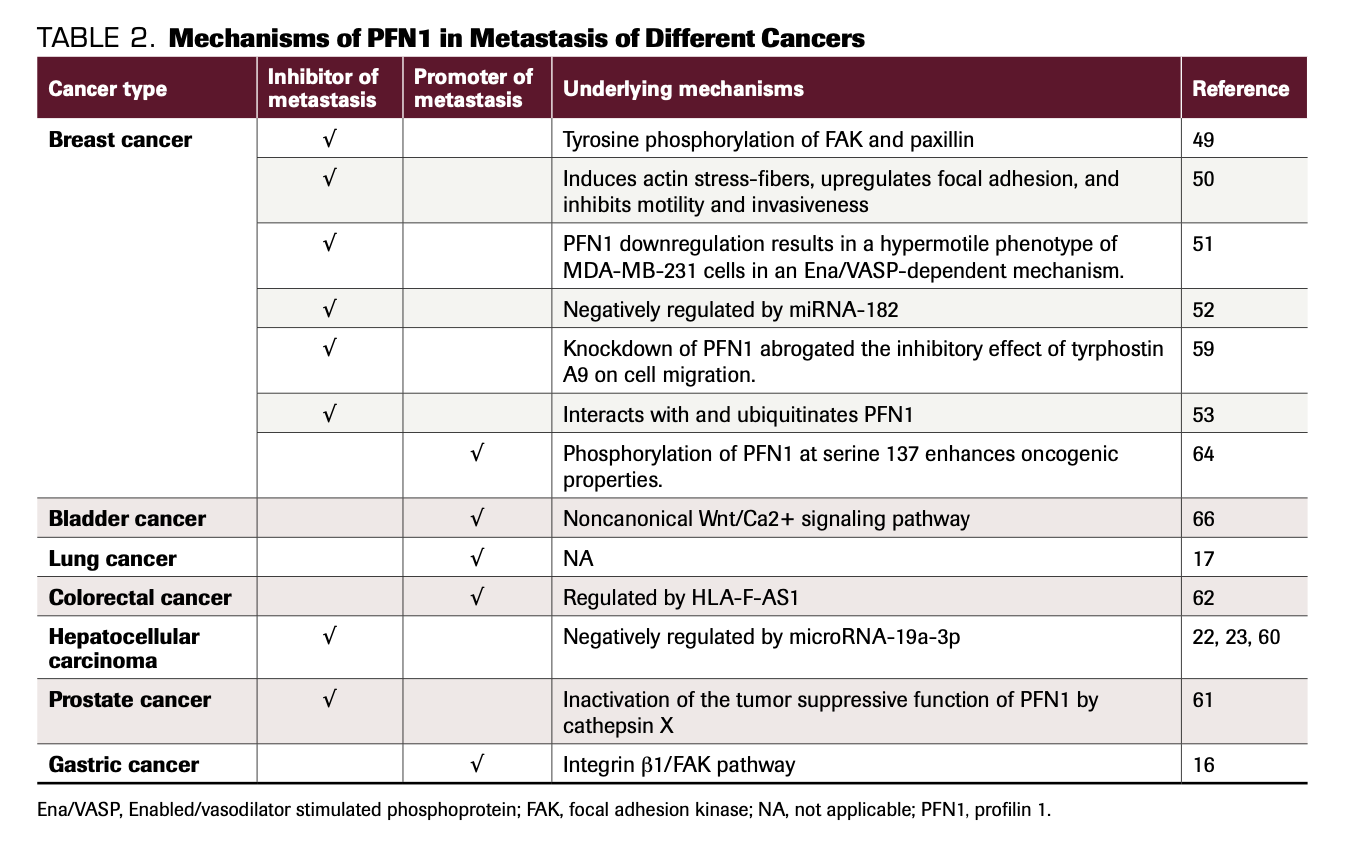
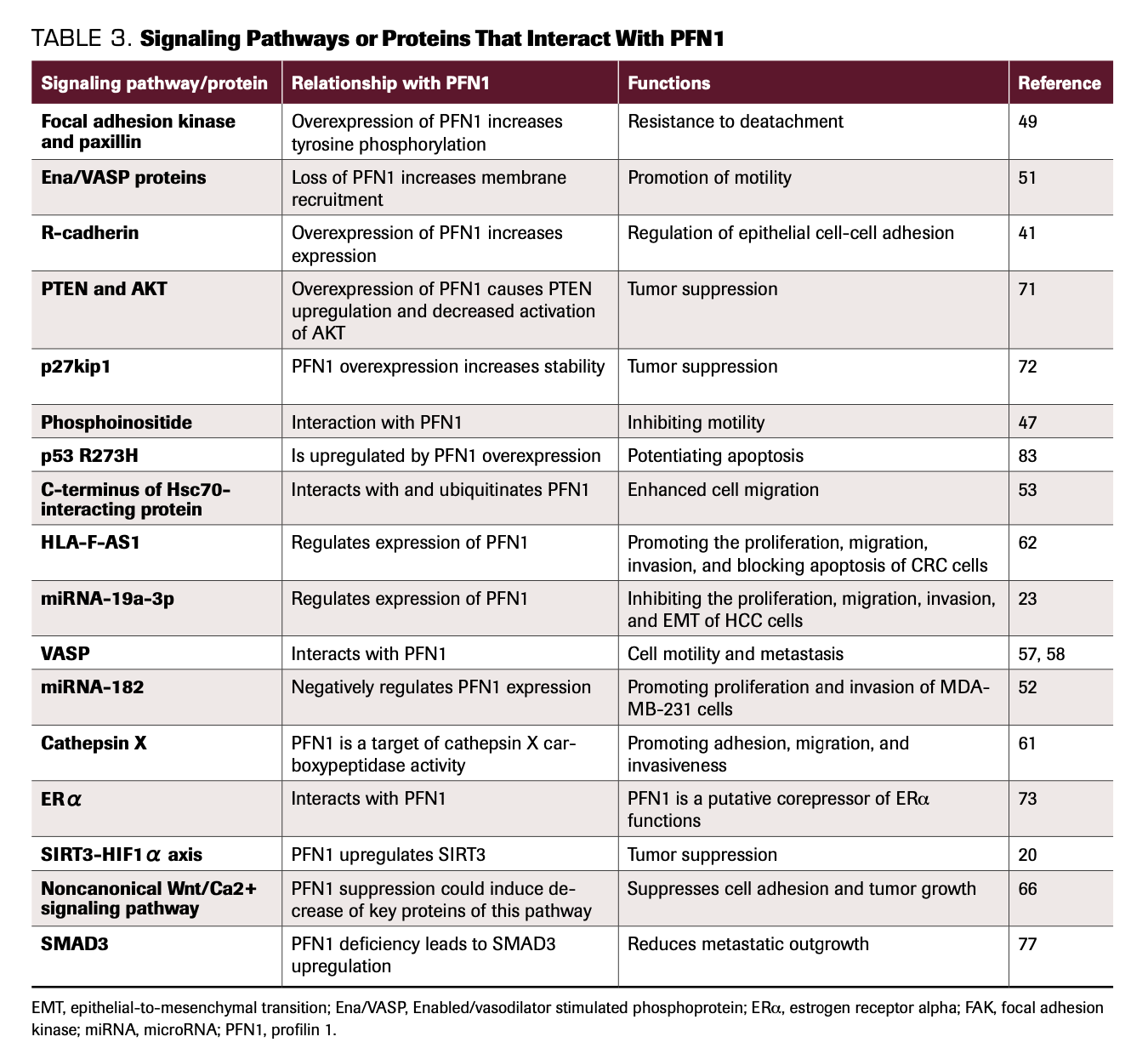
TABLE 3. Signaling Pathways or Proteins That Interact With PFN1
PFN1 as a regulator of proliferation, apoptosis, and drug resistance in cancer
In yeast, the gene encoding PFN1 is essential for cytokinesis.67 Early studies revealed that PFN1–/– embryos died as early as the 2-cell stage, while PFN1–/+ embryos displayed reduced survival during embryogenesis compared with wild-type embryos; this indicates that PFN1 is essential for cell division and survival during embryogenesis.68 PFN1 silencing in endothelial cells inhibits proliferation.69 In addition, homozygous deletion of PFN1 in chondrocytes failed to complete abscission at late-stage cytokinesis.70 The results of all these studies imply that PFN1 plays a role in cell proliferation. In breast cancer, PFN1 overexpression (PFN1-OE) has been shown to inhibit cell growth and exert an inhibitory effect on tumorigenesis,25,40,52,71-75 and PFN1-OE suppresses the activation of AKT, which in turn inhibits the growth of tumor cells.71 PFN1-OE cells arrested at the G1 phase, which was partly attributed to the upregulation of P27kip1.72 miRNA-182 could downregulate PFN1 expression and promote triple-negative breast cancer cell proliferation.52 However, Yap et al put forward opposite views. The authors’ research results revealed that silencing PFN1 resulted in a multinucleation phenotype of breast cancer cells, thus inhibiting proliferation.76 Recent studies from Chakraborty et al also reported that PFN1 knockdown could upregulate SMAD3 and inhibit the proliferation of breast cancer.77 Results of single-cell studies on the extracellular matrix revealed that stiff extracellular matrix led to upregulation of PFN1, possibly promoting the proliferation of breast cancer.78 Apart from breast cancer, PFN1 was also found to suppress proliferation in pancreatic adenocarcinoma,20 endometrial cancer,79 and HCC.23,60 In gastric cancer, silencing PFN1 caused cell cycle arrest at G0/G1 phase, thus restraining cell proliferation.16 Knockdown of PFN1 could also inhibit the proliferation of laryngeal cancer.18 Our previous studies found that overexpression of PFN1 could promote the proliferation of multiple myeloma cells by accelerating the cell cycle from G1 to S phase.80 PFN1 is indispensable for cytokinesis. Nevertheless, PFN1 is involved in regulating cell proliferation not only by impacting cytokinesis but also by modulating cell cycle–related proteins. Otherwise, PFN1 could also interact with cell signaling pathways and indirectly influence cell proliferation.
Tumor growth is not only about uncontrolled proliferation but also resistance to apoptosis.81 Actin dynamics have notable impacts on multiple stages of apoptosis.82 PFN1, as a critical actin-binding protein, is an indispensable regulator of actin dynamics, through which PFN1 participates in regulating apoptosis. PFN1 overexpression could upregulate the most common tumor-associated hotspot mutation of p53—p53R273H—thus sensitizing cancer cells to apoptosis via the intrinsic apoptotic pathway.83 PFN1 has been shown to facilitate apoptosis of breast cancer cells, thus exerting a suppressive effect on tumorigenesis.73,75,83,84 By inducing apoptosis and reducing autophagy, PFN1 has also been shown to sensitize pancreatic cancer cells to irradiation. Additionally, overexpression of PFN1 can significantly elevate apoptotic markers such as cleaved caspase-3 and cleaved PARP after irradiation, suggesting that PFN1 can modulate radiosensitivity partly by regulating apoptosis.85
Given that PFN1 is involved in cell proliferation and apoptosis, it is not difficult to understand its roles in the drug resistance of tumor cells. PFN1 was found to be downregulated in butyrate-treated CRC cells,86 and proteomics studies revealed that PFN1 was differentially expressed in erinacine A–treated CRC cells,87 which suggested the roles of PFN1 in drug-mediated cell death and inhibition of proliferation. In addition, proteomics showed that PFN1 was differentially expressed in mitotane-treated adrenocortical carcinoma,88 and PFN1 was found to be increased in tocotrienol-treated MDA-MB-231 cells,89 indicating its roles in predicting the response to anticancer therapies. Compared with temozolomide (TMZ)-treated glioblastoma cells, PFN1 was downregulated in OKN-007 combined with TMZ-treated glioblastoma cells. Further study results revealed that PFN1 is involved in TMZ resistance.90 Results of our previous studies showed that PFN1 could interact with the Beclin 1 complex and participate in bortezomib resistance in multiple myeloma.80 Since PFN1 is involved in multiple cell processes, including proliferation, apoptosis, and proteomics, it was recognized as a biomarker for therapy sensitivity, and it is worth further exploring its roles in drug resistance. In addition, PFN1 was found to participate in angiogenesis,91-92 initiation of tumors,93 and autophagy.80 Loss of PFN1 in A549 cell lines resulted in fewer early apoptotic cells after treatment with piperlongumine, and PFN1 sensitized A549 cells to anticancer agents.17 PFN1 serves as a bridge for actin-cytoskeleton and cell signaling pathways and is involved in multiple biological and physiological processes. Dysregulation of PFN1 in cancer cells has a notable impact on sensitivity to chemotherapy or radiotherapy and may be a new target for the treatment of drug-resistant or radioresistant patients.
Other functions of PFN1 in tumors
Studies have already confirmed that PFN1 is essential for cell survival in early embryos, as PFN1-KN could induce Drosophila embryos to die at the 2-cell stage.94 For further investigation of PFN1’s roles in tissue-specific stem cells, Zheng et al established PFN1flox/flox mice that inducibly delete PFN1 in HSCs. Results showed that PFN1 was essential for the retention and metabolism of mouse hematopoietic stem cells in bone marrow partially through the axis of PFN1/Gα13/EGR1.95 These study results implied important roles of PFN1 in stem cell function, which were still unclear and deserved further research. Later study results have found that both overexpression and depletion of PFN1 could reduce the stem-like phenotype of MDA-MB-231 (MDA-231) triple-negative breast cancer cells, suggesting that a balanced expression of PFN1 was required for maintenance of optimal stemness and tumor-initiating ability of breast cancer cells.93 Considering that tumor heterogeneity is still an ongoing challenge for cancer treatment and that cancer stem cells (CSC) are considered to be a determining factor of tumor heterogeneity,96 intensive studies on PFN1’s roles in CSC may provide us new insight into tumor initiation.
As mentioned above, PFN1 has been shown to be a critical participator of actin dynamics and to play important roles in cell migration. For cytotoxic T lymphocytes (CTLs), migration abilities are essential for patrolling tissues and locating targeted cells.97-98 Schoppmeyer et al thus studied PFN1’s roles in CTL functions. The authors found that PFN1 negatively regulated CTL-mediated elimination of target cells and that PFN1 downregulation promoted CTL invasion into a 3D matrix in vitro. In patients with pancreatic cancer, PFN1 expression was substantially decreased in peripheral CD8+ T cells.99 However, considering the complexity of immune responses in vivo, the exact roles of PFN1 in tumor immunity remain unclear and need to be further explored.
Conclusions
Based on previous studies, we found that PFN1participates in multiple biological processes of tumor development and progression. Meanwhile, it is noteworthy that PFN1 plays opposite roles in different tumors and at different periods of tumor, potentially leading to the conclusion that PFN1’s function in tumor has spatial and temporal specificity. Future studies on PFN1 should take this into account. PFN1 was shown to be of great significance for diagnosis and prognosis prediction and for monitoring the therapeutic effect of anticancer drugs, and PFN1’s roles in tumor stemness and immunity may provide a new avenue for cancer therapy. Although much research has been done on PFN1 and cancer, puzzles still need to be solved. With deepening research, the function of PFN1 in cancer would be further clarified and its clinical value would be more prominent.
Financial Disclosure: The authors have no significant financial interest in or other relationship with the manufacturer of any product or provider of any service mentioned in this article.
Conflicts of Interest: Authors declare no conflicts of interest for this article.
Acknowledgment: The authors are thankful for financial support from the Doctoral Fund Project of Hunan Provincial People’s Hospital (program number BSJJ201812).
References
1. Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14(8):461-469. doi:10.1016/j.tcb.2004.07.003
2. Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115(3):465-483. doi:10.1016/0022-2836(77)90166-8
3. Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosin beta 4 and profilin: a potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell. 1992;3(9):1015-1024. doi:10.1091/mbc.3.9.1015
4. Yarmola EG, Bubb MR. How depolymerization can promote polymerization: the case of actin and profilin. BioEssays. 2009;31(11):1150-1160. doi:10.1002/bies.200900049
5. Boukhelifa M, Moza M, Johansson T, et al. The proline-rich protein palladin is a binding partner for profilin. FEBS J. 2006;273(1):26-33. doi:10.1111/j.1742-4658.2005.05036.x
6. Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314(6010):472-474. doi:10.1038/314472a0
7. Wu C-H, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488(7412):499-503. doi:10.1038/nature11280
8. Hassona MD, Abouelnaga ZA, Elnakish MT, et al. Vascular hypertrophy-associated hypertension of profilin1 transgenic mouse model leads to functional remodeling of peripheral arteries. Am J Physiol Heart Circ Physiol. 2010;298(6):H2112-H2120. doi:10.1152/ajpheart.00016.2010
9. Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369(9574):1742-1757. doi:10.1016/S0140-6736(07)60781-8
10. Cifani P, Kentsis A. Towards comprehensive and quantitative proteomics for diagnosis and therapy of human disease. Proteomics. 2017;17(1-2):10.1002/pmic.201600079. doi:10.1002/pmic.201600079
11. Masui O, White MA, DeSouza LV, et al. Quantitative proteomic analysis in metastatic renal cell carcinoma reveals a unique set of proteins with potential prognostic significance. Mol Cell Proteomics. 2013;12(1):132-144. doi:10.1074/mcp.M112.020701
12. Neely BA, Wilkins CE, Marlow LA, et al. Proteotranscriptomic analysis reveals stage specific changes in the molecular landscape of clear-cell renal cell carcinoma. PLoS One. 2016;11(4):e0154074.doi:10.1371/journal.pone.0154074
13. Allen A, Gau D, Francoeur P, et al. Actin-binding protein profilin1 promotes aggressiveness of clear-cell renal cell carcinoma cells. J Biol Chem. 2020;295(46):1563615649. doi:10.1074/jbc.RA120.013963
14. Karamchandani JR, Gabril MY, Ibrahim R, et al. Profilin-1 expression is associated with high grade and stage and decreased disease-free survival in renal cell carcinoma. Hum Pathol. 2014;46(5):673-680. doi:10.1016/j.humpath.2014.11.007
15. Tanaka M, Sasaki H, Kino I, Sugimura T, Terada M. Genes preferentially expressed in embryo stomach are predominantly expressed in gastric cancer. Cancer Res. 1992;52(12):3372-3377.
16. Cheng Y-J, Zhu Z-X, Zhou J-S, et al. Silencing profilin-1 inhibits gastric cancer progression via integrin β1/focal adhesion kinase pathway modulation. World J Gastroenterol. 2015;21(8):2323-2335.doi:10.3748/wjg.v21.i8.2323
17. Gagat M, Hałas-Wiśniewska M, Zielińska W, Izdebska M, Grzanka D, Grzanka A. The effect of piperlongumine on endothelial and lung adenocarcinoma cells with regulated expression of profilin-1. Onco Targets Ther. 2018;11:8275-8292. doi:10.2147/OTT.S183191
18. Li L, Zhang Z, Wang C, et al. Quantitative proteomics approach to screening of potential diagnostic and therapeutic targets for laryngeal carcinoma. PLoS One. 2014;9(2):e90181. doi:10.1371/journal.pone.0090181
19. Liang J-W, Shi Z-Z, Shen T-Y, et al. Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization. PLoS One. 2014;9(12):e114616. doi:10.1371/journal.pone.0114616
20. Yao WT, Ji SR, Qin Y, et al. Profilin-1 suppresses tumorigenicity in pancreatic cancer through regulation of the SIRT3-HIF1α axis. Mol Cancer. 2014;13:187. doi:10.1186/1476-4598-13-187
21. Adami GR, O’Callaghan TN, Kolokythas A, Cabay RJ, Zhou Y, Schwartz JL. A loss of profilin-1 in late-stage oral squamous cell carcinoma. J Oral Pathol Med. 2017;46(7):489-495. doi:10.1111/jop.12523
22. Shen K, Xi ZC, Xie JL, et al. Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin1. Oncotarget. 2016;7(35):56650-56663.doi:10.18632/oncotarget.10992
23. Wang Z, Shi Z, Zhang L, Zhang H, Zhang Y. Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in human hepatocellular carcinoma. Pathol Res Pract. 2019;215(3):499-505. doi:10.1016/j.prp.2018.12.012
24. Chan CML, Wong SCC, Lam MYY, et al. Proteomic comparison of nasopharyngeal cancer cell lines C666-1 and NP69 identifies down-regulation of annexin II and beta2-tubulin for nasopharyngeal carcinoma. Arch Pathol Lab Med. 2008;132(4):675-683. doi:10.1043/1543-2165(2008)132675:PCONCC]2.0.CO;2
25. Janke J, Schlüter K, Jandrig B, et al. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J Exp Med. 2000;191(10):1675-1686. doi:10.1084/jem.191.10.1675
26. Peng Z-M, Yu W, Xie Y, et al. A four actin-binding protein signature model for poor prognosis of patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(9):5950-5959.
27. Ding Z, Bae YH, Roy P. Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adh Migr. 2012;6(5):442-449. doi:10.4161/cam.21832
28. Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2019;19(1):11. doi:10.1039/c8lc00684a
29. Zhang P-J, Wei R, Wen X-Y, et al. Genes expression profiling of peripheral blood cells of patients with hepatocellular carcinoma. Cell Biol Int. 2012;36(9):803-809. doi:10.1042/CBI20100920
30. Xie H, Xue Y-Q, Liu P, et al. Multi-parameter gene expression profiling of peripheral blood for early detection of hepatocellular carcinoma. World J Gastroenterol. 2018;24(3):371-378. doi:10.3748/wjg.v24.i3.371
31. Tan Y, Ma S-Y, Wang F-Q, et al. Proteomic-based analysis for identification of potential serum biomarkers in gallbladder cancer. Oncol Rep. 2011;26(4):853-859. doi:10.3892/or.2011.1353
32. Zoidakis J, Makridakis M, Zerefos PG, et al. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol Cell Proteomics. 2012;11(4):M111.009449. doi:10.1074/mcp.M111.009449
33. Grønborg M, Zakarias Kristiansen T, Iwahori A, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157-171. doi:10.1074/mcp.M500178-MCP200
34. Minamida S, Iwamura M, Kodera Y, et al. Profilin 1 overexpression in renal cell carcinoma. Int J Urol. 2011;18(1):63-71. doi:10.1111/j.1442-2042.2010.02670.x
35. Braun M, Fountoulakis M, Papadopoulou A, et al. Down-regulation of microfilamental network-associated proteins in leukocytes of breast cancer patients: potential application to predictive diagnosis. Cancer Genomics Proteomics. 2009;6(1):31-40.
36. Ji H, Greening DW, Kapp EA, Moritz RL, impson RJ. Secretome-based proteomics reveals sulindac-modulated proteins released from colon cancer cells. Proteomics Clin Appl. 2009;3(4):433-451.doi:10.1002/prca.200800077
37. Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73(12):2291-2305. doi:10.1016/j.jprot.2010.07.001
38. Pavlou MP, Diamandis EP. The cancer cell secretome: a good source for discovering biomarkers? J Proteomics. 2010;73(10):1896-1906. doi:10.1016/j.jprot.2010.04.003
39. Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol.2002;12(3):112-120. doi:10.1016/s0962-8924(01)02237-1
40. Wittenmayer N, Jandrig B, Rothkegel M, et al. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15(4):1600-1608. doi:10.1091/mbc.e03-12-0873
41. Zou L, Hazan R, Roy P. Profilin-1 overexpression restores adherens junctions in MDA-MB-231 breast cancer cells in R-cadherin-dependent manner. Cell Motil Cytoskeleton. 2009;66(12):1048-1056. doi:10.1002/cm.20407
42. Jiang C, Veon W, Li H, Hallows KR, Roy P. Epithelial morphological reversion drives Profilin-1-induced elevation of p27(kip1) in mesenchymal triple-negative human breast cancer cells through AMP-activated protein kinase activation. Cell Cycle. 2015;14(18):2914-2923. doi:10.1080/15384101.2015.1069929
43. Beaty BT, Wang Y, Bravo-Cordero JJ, et al. Talin regulates moesin–NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J Cell Biol. 2014;205(5):737-751. doi:10.1083/jcb.201312046
44. Beaty BT, Sharma VP, Bravo-Cordero JJ, et al. β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol Biol Cell. 2013;24(11):1661-1675,S1-S11. doi:10.1091/mbc.E12-12-0908
45. Mader CC, Oser M, Magalhaes MAO, et al. An EGFR–Src–Arg–cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71(5):1730-1741. doi:10.1158/0008-5472.CAN-10-1432
46. Oser M, Yamaguchi H, Mader CC, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186(4):571-587. doi:10.1083/jcb.200812176
47. Baea YH, Dinga ZJ, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci U S A.2010;107(50):21547-21552. doi:10.1073/pnas.1002309107
48. Valenzuela-Iglesias A, Sharma VP, Beaty BT, et al. Profilin1 regulates invadopodium maturation in human breast cancer cells. Eur J Cell Biol. 2015;94(2):78-89. doi:10.1016/j.ejcb.2014.12.002
49. Roy P, Jacobson K. Overexpression of profilin reduces the migration of invasive breast cancer cells. Cell Motil Cytoskeleton. 2004;57(2):84-95. doi:10.1002/cm.10160
50. Zou L, Jaramillo M, Whaley D, et al. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer. 2007;97(10):1361-1371. doi:10.1038/sj.bjc.6604038
51. Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219(2):354-364. doi:10.1002/jcp.21677
52. Liu H, Wang Y, Li X, et al. Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumour Biol. 2013;34(3):1713-1722. doi:10.1007/s13277-013-0708-0
53. Choi YN, Lee SK, Seo TW, Lee JS, Yoo SJ. C-terminus of Hsc70-interacting protein regulates profilin1 and breast cancer cell migration. Biochem Biophys Res Commun. 2014;446(4):1060-1066.doi:10.1016/j.bbrc.2014.03.061
54. Ding Z, Joy M, Bhargava R, et al. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene. 2014;33(16):2065-2074. doi:10.1038/onc.2013.166
55. Mouneimne G, Hansen SD, Selfors LM, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22(5):615-630.doi:10.1016/j.ccr.2012.09.027
56. Liu Y, Wu C, Zhu T, Sun W. LMO2 enhances lamellipodia/filopodia formation in basal-type breast cancer cells by mediating ARP3-profilin1 interaction. Med Sci Monit. 2017;23:695-703. doi:10.12659/msm.903261
57. Gau D, Veon W, Shroff SG, Roy P. The VASP–profilin1 (Pfn1) interaction is critical for efficient cell migration and is regulated by cell–substrate adhesion in a PKA-dependent manner. J Biol Chem.2019;294(17):6972-6985. doi:10.1074/jbc.RA118.005255
58. Gau D, Ding ZJ, Baty C, Roy P. Fluorescence resonance energy transfer (FRET)-based detection of profilin–VASP interaction. Cell Mol Bioeng. 2011;4(1):1-8. doi:10.1007/s12195-010-0133-z
59. Joy ME, Vollmer LL, Hulkower K, et al. A high-content, multiplexed screen in human breast cancer cells identifies profilin-1 inducers with anti-migratory activities. PLoS One. 2014;9(2):e88350.doi:10.1371/journal.pone.0088350
60. Wu N, Zhang W, Yang Y, et al. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid–treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics.2006;6(22):6095-6106. doi:10.1002/pmic.200500321
61. Pečar Fonović U, Jevnikar Z, Rojnik M, et al. Profilin 1 as a target for cathepsin X activity in tumor cells. PLoS One. 2013;8(1):e53918. doi:10.1371/journal.pone.0053918
62. Huang Y, Sun H, Ma X, et al. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 2019;254:117180. doi:10.1016/j.lfs.2019.117180
63. Bai N, Ma Y, Zhao J, Li B. Knockdown of lncRNA HCP5 suppresses the progression of colorectal cancer by miR-299-3p/PFN1/AKT axis. Cancer Manag Res. 2020;12:4747-4758. doi:10.2147/CMAR.S255866
64. Rizwani W, Fasim A, Sharma D, Reddy DJ, Bin Omar NAM, Singh SS. S137 phosphorylation of profilin 1 is an important signaling event in breast cancer progression. PLoS One. 2014;9(8):e103868.doi:10.1371/journal.pone.0103868
65. Ma Y, Li Y-F, Wang T, Pang R, Xue Y-W, Zhao S-P. Identification of proteins associated with lymph node metastasis of gastric cancer. J Cancer Res Clin Oncol. 2014;140(10):1739-1749. doi:10.1007/s00432-014-1679-2
66. Frantzi M, Klimou Z, Makridakis M, et al. Silencing of Profilin-1 suppresses cell adhesion and tumor growth via predicted alterations in integrin and Ca2+ signaling in T24M-based bladder cancer models. Oncotarget.2016;7(43):70750-70758. doi:10.18632/oncotarget.12218
67. Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125(6):1289-1301. doi:10.1083/jcb.125.6.1289
68. Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A. 2001;98(7):3832-3836.doi:10.1073/pnas.051515498
69. Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119(Pt 19):4127-4137. doi:10.1242/jcs.03178
70. Böttcher RT, Wiesner S, Braun A, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 2009;28(8):1157-1169. doi:10.1038/emboj.2009.58
71. Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218(2):436-443. doi:10.1002/jcp.21618
72. Zou L, Ding Z, Roy P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J Cell Physiol. 2010;223(3):623-629. doi:10.1002/jcp.22058
73. Kumar Kanaujiya J, Lochab S, Kapoor I, et al. Proteomic identification of Profilin1 as a corepressor of estrogen receptor alpha in MCF7 breast cancer cells. Proteomics. 2013;13(14):2100-2112.doi:10.1002/pmic.201200534
74. Coumans JVF, Gau D, Poljak A, Wasinger V, Roy P, Moens PDJ. Profilin-1 overexpression in MDA-MB-231 breast cancer cells is associated with alterations in proteomics biomarkers of cell proliferation, survival, and motility as revealed by global proteomics analyses. OMICS. 2014;18(12):779-791. doi:10.1089/omi.2014.0075
75. Diamond MI, Cai S, Boudreau A, et al. Subcellular localization and Ser-137 phosphorylation regulate tumor-suppressive activity of profilin-1. J Biol Chem. 2015;290(14):9075-9086. doi:10.1074/jbc.M114.619874
76. Yap KL, Fraley SI, Thiaville MM, et al. NAC1 is an actin-binding protein that is essential for effective cytokinesis in cancer cells. Cancer Res. 2012;72(16):4085-4096. doi:10.1158/0008-5472.CAN-12-0302
77. Chakraborty S, Jiang C, Gau D, et al. Profilin-1 deficiency leads to SMAD3 upregulation and impaired 3D outgrowth of breast cancer cells. Br J Cancer. 2018;119(9):1106-1117. doi:10.1038/s41416-018-0284-6
78. Sun YP, Deng RJ, Zhang KX, Ren XJ, Zhang L, Li JH. Single-cell study of the extracellular matrix effect on cell growth by in situ imaging of gene expression. Chem Sci. 2017;8(1):8019-8024. doi:10.1039/c7sc03880a
79. George L, Winship A, Sorby K, Dimitriadis E, Menkhorst E. Profilin-1 is dysregulated in endometroid (type I) endometrial cancer promoting cell proliferation and inhibiting pro-inflammatory cytokine production. Biochem Biophys Res Commun. 2020;531(4):459-464. doi:10.1016/j.bbrc.2020.07.123
80. Lu Y, Wang Y, Xu H, Shi C, Jin F, Li W. Profilin 1 induces drug resistance through Beclin1 complex–mediated autophagy in multiple myeloma. Cancer Sci. 2018;109(9):2706-2716. doi:10.1111/cas.13711
81. Wu J. Apoptosis and angiogenesis: two promising tumor markers in breast cancer (review). Anticancer Res. 1996;16(4B):2233-2239.
82. Franklin-Tong VE, Gourlay CW. A role for actin in regulating apoptosis/programmed cell death: evidence spanning yeast, plants and animals. Biochem J. 2008;413(3):389-404. doi:10.1042/BJ20080320
83. Yao W, Cai X, Liu C, et al. Profilin 1 potentiates apoptosis induced by staurosporine in cancer cells. Curr Mol Med. 2013;13(3):417-428.
84. Yao W, Yu X, Fang Z, et al. Profilin1 facilitates staurosporine-triggered apoptosis by stabilizing the integrin β1–actin complex in breast cancer cells. J Cell Mol Med. 2012;16(4):824-835. doi:10.1111/j.1582-4934.2011.01369.x
85. Cheng H, Li J, Liu C, et al. Profilin1 sensitizes pancreatic cancer cells to irradiation by inducing apoptosis and reducing autophagy. Curr Mol Med. 2013;13(8):1368-1375. doi:10.2174/15665240113139990060
86. Fung KYC, Cursaro C, Lewanowitsch T, et al. A combined free-flow electrophoresis and DIGE approach to identify proteins regulated by butyrate in HT29 cells. Proteomics. 2011;11(5):964-971.doi:10.1002/pmic.201000429
87. Lee K-C, Kuo H-C, Shen C-H, et al. A proteomics approach to identifying novel protein targets involved in erinacine A–mediated inhibition of colorectal cancer cells’ aggressiveness. J Cell Mol Med. 2017;21(3):588-599. doi:10.1111/jcmm.13004
88. Stigliano A, Cerquetti L, Borro M, et al. Modulation of proteomic profile in H295R adrenocortical cell line induced by mitotane. Endocr Relat Cancer. 2008;15(1):1-10. doi:10.1677/ERC-07-0003
89. Ramdas P, Radhakrishnan AK, Abdu Sani AA, Abdul-Rahman PS. Tocotrienols modulate breast cancer secretomes and affect cancer-signaling pathways in MDA-MB-231 cells: a label-free quantitative proteomic analysis. Nutr Cancer. 2019;71(8):1263-1271. doi:10.1080/01635581.2019.1607407
90. Towner RA, Smith N, Saunders D, et al. OKN-007 increases temozolomide (TMZ) sensitivity and suppresses TMZ-resistant glioblastoma (GBM) tumor growth. Transl Oncol. 2019;12(2):320-335. doi:10.1016/j.tranon.2018.10.002
91. Fan Y, Potdar AA, Gong Y, et al. Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat Cell Biol. 2014;16(5):445-456. doi:10.1038/ncb2954
92. Fan Y, Arif A, Gong YQ, et al. Stimulus-dependent phosphorylation of profilin-1 in angiogenesis. Nat Cell Biol. 2012;14(10):1046-1056. doi:10.1038/ncb2580
93. Jiang C, Ding Z, Joy M, et al. A balanced level of profilin-1 promotes stemness and tumor-initiating potential of breast cancer cells. Cell Cycle. 2017;16(24):2366-2373. doi:10.1080/15384101.2017.1346759
94. Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120(4):717-728.
95. Zheng J, Lu Z, Kocabas F, et al. Profilin 1 is essential for retention and metabolism of mouse hematopoietic stem cells in bone marrow. Blood. 2014;123(7):992-1001. doi:10.1182/blood-2013-04-498469
96. Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer.2017;16(1):41. doi:10.1186/s12943-017-0600-4
97. Weigelin B, Krause M, Friedl P. Cytotoxic T lymphocyte migration and effector function in the tumor microenvironment. Immunol Lett. 2011;138(1):19-21. doi:10.1016/j.imlet.2011.02.016
98. Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nature Immunol. 2008;9(9):960-969. doi:10.1038/ni.f.212
99. Schoppmeyer R, Zhao R, Cheng H, et al. Human profilin 1 is a negative regulator of CTL mediated cell-killing and migration. Eur J Immunol. 2017;47(9):1562-1572. doi:10.1002/eji.201747124
